













| General Information: |
|
| Name(s) found: |
RRD2 /
YPL152W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 358 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[TAS cell fraction [IDA |
| Biological Process: |
response to osmotic stress
[IGI mitotic spindle organization in nucleus [IMP |
| Molecular Function: |
protein phosphatase type 2A regulator activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLPEKRLLTP DDMKLWEESP TRAHFTKFII DLAESVKGHE NSQYKEPISE SINSMMNLLS 60
61 QIKDITQKHP VIKDADSSRF GKVEFRDFYD EVSRNSRKIL RSEFPSLTDE QLEQLSIYLD 120
121 ESWGNKRRID YGSGHELNFM CLLYGLYSYG IFNLSNDSTN LVLKVFIEYL KIMRILETKY 180
181 WLEPAGSHGV WGLDDYHFLP FLFGAFQLTT HKHLKPISIH NNELVEMFAH RYLYFGCIAF 240
241 INKVKSSASL RWHSPMLDDI SGVKTWSKVA EGMIKMYKAE VLSKLPIMQH FYFSEFLPCP 300
301 DGVSPPRGHI HDGTDKDDEC NFEGHVHTTW GDCCGIKLPS AIAATEMNKK HHKPIPFD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CDC55 PPH22 RRD2 RTS1 TPD3 ZDS1 |
| View Details | Krogan NJ, et al. (2006) |

|
CDC55 FIR1 PHR1 PPH21 PPH22 RRD2 TPD3 YRR1 ZDS1 ZDS2 |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC55 CIN1 ERG13 GUS1 HHF1, HHF2 HOS2 HOS4 IML1 KAP95 KEL1 LTE1 MYO5 PFK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RRD2 RTS1 RTS3 SCL1 SET3 SIF2 SNT1 SRP1 TDH2 TDH3 TEF4 TPD3 YBL104C YOR1 YRA1 ZDS1 ZDS2 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
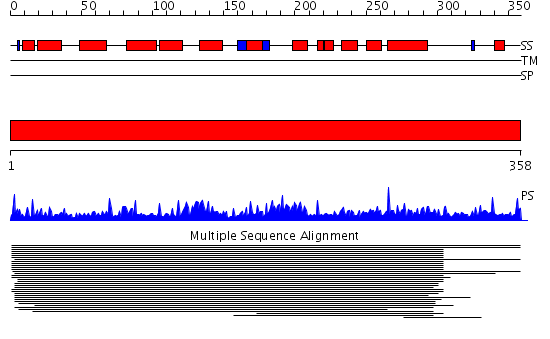
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..358] | 194.744727 | Phosphotyrosyl phosphate activator (PTPA) protein No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)