













| General Information: |
|
| Name(s) found: |
RTT101 /
YJL047C
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 842 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
regulation of mitosis
[IMP negative regulation of transposition, DNA-mediated [IMP ubiquitin-dependent protein catabolic process [IC |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA protein binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MINESVSKRE GFHESISRET SASNALGLYN KFNDERNPRY RTMIAELHEF FHLTLAETIT 60
61 ETDVKELECN KEKAAKFRKL MPKMLNNCRE LTQRKSYIPY NSEFNGNDEK QKKFQLLHQH 120
121 QIVLSFQEFC DELAKLIIDA HVLSFLTRCD YSYEIIPKNW TSFYKLFQYV MGAVGPIISY 180
181 VPVNYPMIRK ELGFETLTIF QYYDSKLFEC MKSHFGREFS TLVSATIHHY IHMFPITNTM 240
241 LEKEVPMLRI MSNCNFSIEG LSPKDFYMKT LRQYYCEESN LRPRLETFKN FKVLLTRNAL 300
301 LASLFSPEWV SDANDLFISH LLLNKKSISE YIEIGKDTYD EEKERYFKTE THFSLLMFRN 360
361 AFEAKNMLSK FKEFCDDAVS EKLKAAYGSN HDTERLFDEV VQLANVDHLK IYSDSIEYHL 420
421 CNLLGSTSKA IEQYVKYFES HLFIIVRKIK TTKKDLPRDM KIKYLNENLP ILRLKFVNLP 480
481 TFPNFFERSI FRKTILQSDQ NSSFIKDILP VYKDSLMELF KQRIITNVSQ EDEMRYRDQY 540
541 QPYLSQFFQP VEVMADLRIK YASFLSFYEN IEAAVKFGKT YNENNSKSFF PLIFDRERIP 600
601 KVFQQSNEVK KNFVLPQEMD DTWNQFLRNY HEQNKVEDSD ASKKELYPMW NLHHCEVESP 660
661 YIIQDGTNLI FELTLFQTCV LTLFNESDHL TLQVISEQTK LAYKDLALVL KSFCNYKILT 720
721 RDIDNTYSIN ESFKPDMKKV KNGKLRVVLP RTASLQSSNT GGERTSSAHH EGSNSQWTQE 780
781 LLKACITRSV KSERNGLDYD HLFETVKQQI KGFSVGEFKD ALAKLLRDKF ITRDESTATY 840
841 KY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
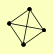
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CDC34 CDC53 RTG2 RTT101 |
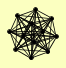
| View Details | Krogan NJ, et al. (2006) |
|
CAK1 CDC34 CDC53 CDC9 CRT10 GRR1 MEP3 MET30 MMS1 POR1 RTT101 SAF1 |
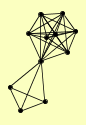
| View Details | Ho Y, et al. (2002) |
|
CDC53 GDH2 GUF1 HRT1 IDH1 MMS22 RIX7 RPN8 RTT101 RTT107 SEC27 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..369] [383..396] |
164.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 | |
| 2 | View Details | [370..382] [397..551] |
164.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 | |
| 3 | View Details | [552..649] | 164.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 | |
| 4 | View Details | [650..773] | 164.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 | |
| 5 | View Details | [774..842] | 164.0 | Anaphase promoting complex (APC); Cullin homolog 1, Cul-1; Cullin homolog 1, cul-1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)