













| General Information: |
|
| Name(s) found: |
SAF1 /
YBR280C
[SGD]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 637 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
proteasomal ubiquitin-dependent protein catabolic process
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEVESREKE PDAGLSPDIV QATLPFLSSD DIKNLSQTNK YYNTLLDFDH SKILWHELFH 60
61 KAFGTLKTND EPFQGRNSAE FKTCTETILR EAFPTLSWQE VYQLRAYDAK FYSWGYLKHG 120
121 RLGYTASSNN ELVATSLNGP SPRFKYGVNT PTEVPWFNSR TTSRTSNFTP SEDPLSAIKK 180
181 DGDEIIAQVS SGGFSFQILT ESGNLYSSGS TFSGGLKGPG PSGSQHDYNP FREMIHNMER 240
241 SYPRITSRSN GSTVNTTGTF SGRRMSGSHP STAYEPGNAT TAQHITIDSG GAPAASPGGS 300
301 HSGVPRTTMP SMGPHENIYS QIEMLERSAN KAVPGNNHIR RMFARNSFPL YSGRDENLGS 360
361 FNDIQFVAVS SGRSHFLAMD TDNNIYSWDS TESDQGVKIE FANLPSRATN PILKIASGWN 420
421 FNCCYIYKVG LVAWKERDAI QKGESFAFAK YEIVPNTNDV NGDSRIVDFA CLQDNCVFFI 480
481 NNNGDKLWKY HNGLNQIVDL NIVGKLCKIN ACFASLVLFT DTHCYTLKVT NGDVDKDSLT 540
541 ELDINENVIS VASGDYHTVA LTERGHLYSW GIESQDCGCL GLGPSEKIVN ELHIGNWEGQ 600
601 RNIRVVKPTK IELPEDYICV SVTAGGWQTG ALIIKKH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
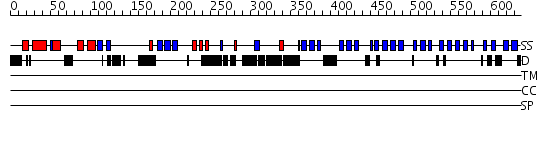
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..267] | 1.05 | Regulator of chromosome condensation RCC1 | |
| 2 | View Details | [268..637] | 60.69897 | Regulator of chromosome condensation RCC1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)