













| General Information: |
|
| Name(s) found: |
RIX7 /
YLL034C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 837 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IDA |
| Biological Process: |
ribosomal large subunit export from nucleus
[IMP |
| Molecular Function: |
ATPase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKVKSKKNS LTSSLDNKIV DLIYRLLEEK TLDRKRSLRQ ESQGEEGENN EGEEDEDIFE 60
61 SMFFAKDLTA GEIFTFCLTK DLSLQRVKKV VLQKTIDRML KDVIESELEE FGSYPGYNNE 120
121 EEEKPSLEEE LAKKNMMIER DTNEMNKRIT STWSKSGSVS ESITETDDPK TEEVKKSKKR 180
181 SKEGTCKVKR QKIKEDRSPP NSSLKSLGGM DDVVAQLMEL IGLPILHPEI FLSTGVEPPR 240
241 GVLLHGPPGC GKTSIANALA GELQVPFISI SAPSVVSGMS GESEKKIRDL FDEARSLAPC 300
301 LVFFDEIDAI TPKRDGGAQR EMERRIVAQL LTSMDELTME KTNGKPVIII GATNRPDSLD 360
361 AALRRAGRFD REICLNVPNE VSRLHILKKM SDNLKIDGAI DFAKLAKLTP GFVGADLKAL 420
421 VTAAGTCAIK RIFQTYANIK STPTTATDSS EDNMEIDETA NGDESSLKNT ANMIDPLPLS 480
481 VVQQFIRNYP EPLSGEQLSL LSIKYEDFLK ALPTIQPTAK REGFATVPDV TWANVGALQR 540
541 VRLELNMAIV QPIKRPELYE KVGISAPGGV LLWGPPGCGK TLLAKAVANE SRANFISIKG 600
601 PELLNKYVGE SERSIRQVFT RARASVPCVI FFDELDALVP RRDTSLSESS SRVVNTLLTE 660
661 LDGLNDRRGI FVIGATNRPD MIDPAMLRPG RLDKSLFIEL PNTEEKLDII KTLTKSHGTP 720
721 LSSDVDFEEI IRNEKCNNFS GADLAALVRE SSVLALKRKF FQSEEIQSVL DNDLDKEFED 780
781 LSVGVSGEEI IVTMSDFRSA LRKIKPSVSD KDRLKYDRLN KKMGLTEEMK DAEEMKQ |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
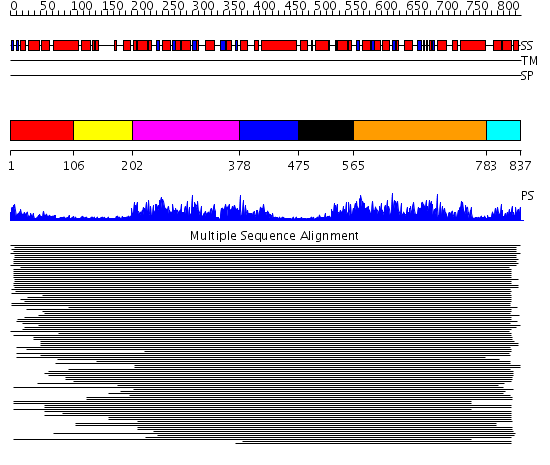
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 630.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 2 | View Details | [106..201] | 630.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [202..377] | 630.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 4 | View Details | [378..474] | 630.9897 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 5 | View Details | [475..564] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [565..782] | 85.886057 | ATPase family associated with various cellular activities (AAA) No confident structure predictions are available. | |
| 7 | View Details | [783..837] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)