













| General Information: |
|
| Name(s) found: |
YTM1 /
YOR272W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 460 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IDA |
| Biological Process: |
ribosomal large subunit biogenesis
[IMP ribosome biogenesis [RCA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEDKSQVKI RFFTREKDEL LHVQDTPMYA PISLKRYGLS EIVNHLLGSE KPVPFDFLIE 60
61 GELLRTSLHD YLTKKGLSSE ASLNVEYTRA ILPPSYLNSF SNEDWVSSLD VGDGSKHIIS 120
121 GSYDGIVRTW DLSGNVQKQY SGHSGPIRAV KYISNTRLVS AGNDRTLRLW KTKNDDLKLT 180
181 SQQQAQEDDD DEVNIEDGKT LAILEGHKAP VVSIDVSDNS RILSASYDNS IGFWSTIYKE 240
241 MTVVDPLEDI NNPNNKISTA ARKRRKLTMK DGTIRRRAPL SLLESHTAPV EQVIFDSTDN 300
301 TVGYSVSQDH TIKTWDLVTA RCIDTRTTSY SLLSIAQLST LNLLACGSSA RHITLHDPRV 360
361 GASSKVTQQQ LIGHKNFVSS LDTCPENEYI LCSGSHDGTV KVWDVRSTSP MYTITREDKS 420
421 VQKGVNDKVF AVKWAEKVGI ISAGQDKKIQ INKGDNIFKN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #28 Asynchronous Prep5-TiO2 Phosphopeptide Enrichment | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
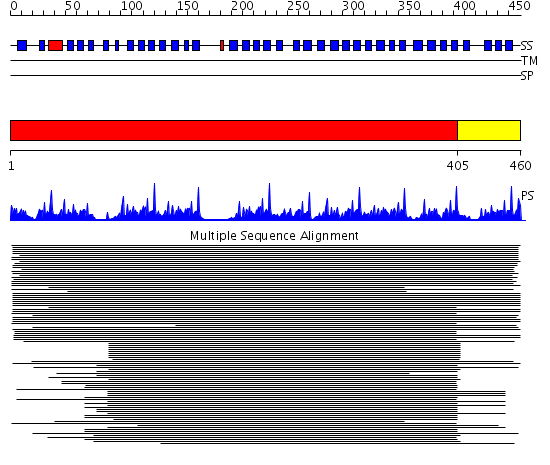
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..404] | 93.06601 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 2 | View Details | [405..460] | 150.0 | Tup1, C-terminal domain | |
| 3 | View Details | [397..460] | 28.522879 | No description for 2pm6B was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)