













| General Information: |
|
| Name(s) found: |
TEM1 /
YML064C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 245 amino acids |
Gene Ontology: |
|
| Cellular Component: |
spindle pole body
[TAS |
| Biological Process: |
regulation of exit from mitosis
[TAS |
| Molecular Function: |
protein binding
[IPI GTPase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATPSTGANN SIPAVRNQVE VQVGLVGDAQ VGKTSLMVKY VQNIYDKEYT QTLGVNFLKR 60
61 KVSIRSTDII FSIMDLGGQR EFINMLPIAT VGSSVIIFLF DLTRPETLSS IKEWYRQAYG 120
121 LNDSAIPILV GTKYDLLIDL DPEYQEQISR TSMKYAQVMN APLIFCSTAK SINIQKIFKI 180
181 ALAKIFNLTL TIPEINEIGD PLLIYKHLGG QQHRHHNKSQ DRKSHNIRKP SSSPSSKAPS 240
241 PGVNT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CDC15 DUG2 TEM1 |
| View Details | Krogan NJ, et al. (2006) |

|
BFA1 CDC48 DOA1 FBP1 MRM1 NDT80 NPL4 OTU1 RDS2 SHP1 TEM1 TUP1 UBX4 UBX5 UBX7 UFD1 UFD2 YDR049W YGL108C YJR098C YNL155W YOR342C |
| View Details | Ho Y, et al. (2002) |

|
ADK1 ATP3 AVO1 BFA1 CCT5 CDC15 CDC33 CLU1 COF1 COR1 CPR1 CPR3 CYS4 DHH1 DPB2 DUG2 DUT1 EFB1 FAA4 FUN12 GAL7 GCD11 GPH1 GUS1 HAS1 IDH1 ILV2 KGD1 MCX1 MET16 MKK2 NMD3 NOG2 NPA3 NUP53 PDX1 PFK1 PHO85 PST2 PUF3 RNR1 RNR2 RPN12 RVB1 SAR1 SEC53 SIS1 SLX1 SRP54 SSD1 STI1 TEF4 TEM1 TFC7 TIF1, TIF2 TMA29 UFD4 VMA5 VMA8 YER077C YGR066C YGR266W YHB1 YHR033W YKR051W YKU80 YLR271W YNK1 YPR003C YTM1 |
| View Details | Qiu et al. (2008) |
|
ATG27 CDC14 HSK3 LCB3 MOB1 TEM1 VRP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
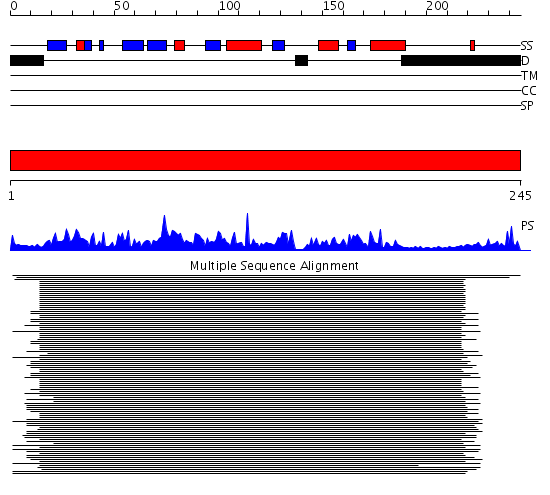
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..245] | 208.522879 | Rab-related protein Sec4 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|