













| General Information: |
|
| Name(s) found: |
UFD4 /
YKL010C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1483 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IPI mitochondrion [IDA |
| Biological Process: |
protein polyubiquitination
[TAS protein monoubiquitination [TAS |
| Molecular Function: |
ubiquitin-protein ligase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSENNSHNLD EHESHSENSD YMMDTQVEDD YDEDGHVQGE YSYYPDEDED EHMLSSVGSF 60
61 EADDGEDDDN DYHHEDDSGL LYGYHRTQNG SDEDRNEEED GLERSHDNNE FGSNPLHLPD 120
121 ILETFAQRLE QRRQTSEGLG QHPVGRTLPE ILSMIGGRME RSAESSARNE RISKLIENTG 180
181 NASEDPYIAM ESLKELSENI LMMNQMVVDR IIPMETLIGN IAAILSDKIL REELELQMQA 240
241 CRCMYNLFEV CPESISIAVD EHVIPILQGK LVEISYIDLA EQVLETVEYI SRVHGRDILK 300
301 TGQLSIYVQF FDFLTIHAQR KAIAIVSNAC SSIRTDDFKT IVEVLPTLKP IFSNATDQPI 360
361 LTRLVNAMYG ICGALHGVDK FETLFSLDLI ERIVQLVSIQ DTPLENKLKC LDILTVLAMS 420
421 SDVLSRELRE KTDIVDMATR SFQHYSKSPN AGLHETLIYV PNSLLISISR FIVVLFPPED 480
481 ERILSADKYT GNSDRGVISN QEKFDSLVQC LIPILVEIYT NAADFDVRRY VLIALLRVVS 540
541 CINNSTAKAI NDQLIKLIGS ILAQKETASN ANGTYSSEAG TLLVGGLSLL DLICKKFSEL 600
601 FFPSIKREGI FDLVKDLSVD FNNIDLKEDG NENISLSDEE GDLHSSIEEC DEGDEEYDYE 660
661 FTDMEIPDSV KPKKISIHIF RTLSLAYIKN KGVNLVNRVL SQMNVEQEAI TEELHQIEGV 720
721 VSILENPSTP DKTEEDWKGI WSVLKKCIFH EDFDVSGFEF TSTGLASSIT KRITSSTVSH 780
781 FILAKSFLEV FEDCIDRFLE ILQSALTRLE NFSIVDCGLH DGGGVSSLAK EIKIKLVYDG 840
841 DASKDNIGTD LSSTIVSVHC IASFTSLNEF LRHRMVRMRF LNSLIPNLTS SSTEADREEE 900
901 ENCLDHMRKK NFDFFYDNEK VDMESTVFGV IFNTFVRRNR DLKTLWDDTH TIKFCKSLEG 960
961 NNRESEAAEE ANEGKKLRDF YKKREFAQVD TGSSADILTL LDFLHSCGVK SDSFINSKLS1020
1021 AKLARQLDEP LVVASGALPD WSLFLTRRFP FLFPFDTRML FLQCTSFGYG RLIQLWKNKS1080
1081 KGSKDLRNDE ALQQLGRITR RKLRISRKTI FATGLKILSK YGSSPDVLEI EYQEEAGTGL1140
1141 GPTLEFYSVV SKYFARKSLN MWRCNSYSYR SEMDVDTTDD YITTLLFPEP LNPFSNNEKV1200
1201 IELFGYLGTF VARSLLDNRI LDFRFSKVFF ELLHRMSTPN VTTVPSDVET CLLMIELVDP1260
1261 LLAKSLKYIV ANKDDNMTLE SLSLTFTVPG NDDIELIPGG CNKSLNSSNV EEYIHGVIDQ1320
1321 ILGKGIEKQL KAFIEGFSKV FSYERMLILF PDELVDIFGR VEEDWSMATL YTNLNAEHGY1380
1381 TMDSSIIHDF ISIISAFGKH ERRLFLQFLT GSPKLPIGGF KSLNPKFTVV LKHAEDGLTA1440
1441 DEYLPSVMTC ANYLKLPKYT SKDIMRSRLC QAIEEGAGAF LLS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
UBC4 UFD4 |
| View Details | Krogan NJ, et al. (2006) |
|
DRS2 MOT2 UBC4 UBC5 UFD4 |
| View Details | Gavin AC, et al. (2006) |

|
AHA1 UFD4 |
| View Details | Ho Y, et al. (2002) |
|
APM1 CDC15 CLU1 DUG2 FET3 HYM1 KRE33 KRE6 PEX19 QCR7 RPN1 TEM1 UBC4 UFD4 YBL104C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
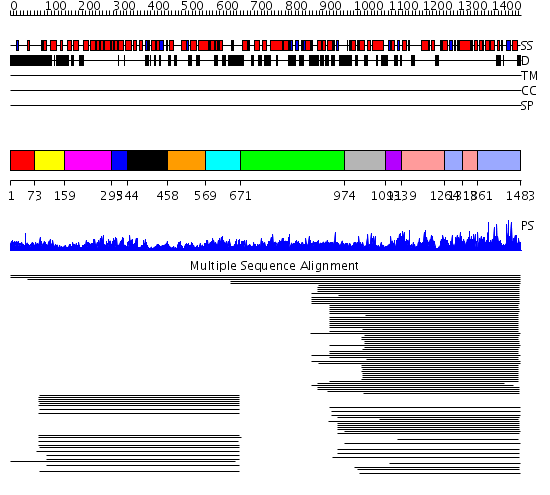
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..72] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [73..158] | 2.09691 | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA | |
| 3 | View Details | [159..294] | 113.0 | Importin alpha | |
| 4 | View Details | [295..343] | 113.0 | Importin alpha | |
| 5 | View Details | [344..457] | 113.0 | Importin alpha | |
| 6 | View Details | [458..568] | 113.0 | Importin alpha | |
| 7 | View Details | [569..670] | 113.0 | Importin alpha | |
| 8 | View Details | [671..973] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [974..1092] | 3.09691 | Ubiquitin ligase NEDD4 WWIII domain | |
| 10 | View Details | [1093..1138] | 323.0103 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 11 | View Details | [1139..1263] [1318..1360] |
323.0103 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 12 | View Details | [1264..1317] [1361..1483] |
323.0103 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)