













| General Information: |
|
| Name(s) found: |
ECI1 /
YLR284C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 280 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA |
| Biological Process: |
fatty acid beta-oxidation
[IDA |
| Molecular Function: |
dodecenoyl-CoA delta-isomerase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQEIRQNEK ISYRIEGPFF IIHLMNPDNL NALEGEDYIY LGELLELADR NRDVYFTIIQ 60
61 SSGRFFSSGA DFKGIAKAQG DDTNKYPSET SKWVSNFVAR NVYVTDAFIK HSKVLICCLN 120
121 GPAIGLSAAL VALCDIVYSI NDKVYLLYPF ANLGLITEGG TTVSLPLKFG TNTTYECLMF 180
181 NKPFKYDIMC ENGFISKNFN MPSSNAEAFN AKVLEELREK VKGLYLPSCL GMKKLLKSNH 240
241 IDAFNKANSV EVNESLKYWV DGEPLKRFRQ LGSKQRKHRL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
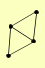
|
DCI1 ECI1 MDH3 POT1 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
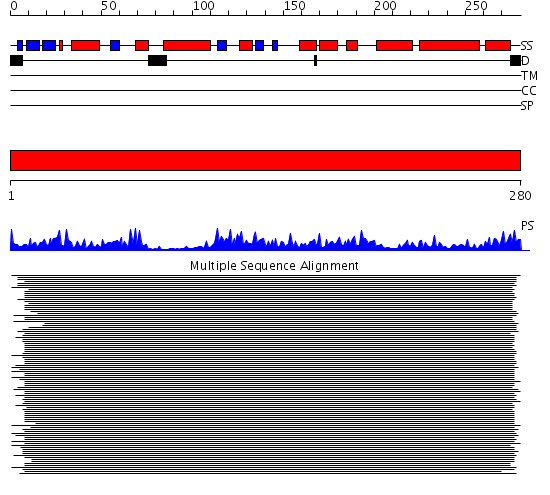
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..280] | 1678.0 | Dienoyl-CoA isomerase (delta3-delta2-enoyl-CoA isomerase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)