













| General Information: |
|
| Name(s) found: |
RET1 /
YOR207C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1149 amino acids |
Gene Ontology: |
|
| Cellular Component: |
DNA-directed RNA polymerase III complex
[TAS |
| Biological Process: |
transcription from RNA polymerase III promoter
[TAS]
|
| Molecular Function: |
DNA-directed RNA polymerase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVAATKRRKT HIHKHVKDEA FDDLLKPVYK GKKLTDEINT AQDKWHLLPA FLKVKGLVKQ 60
61 HLDSFNYFVD TDLKKIIKAN QLILSDVDPE FYLKYVDIRV GKKSSSSTKD YLTPPHECRL 120
121 RDMTYSAPIY VDIEYTRGRN IIMHKDVEIG RMPIMLRSNK CILYDADESK MAKLNECPLD 180
181 PGGYFIVNGT EKVILVQEQL SKNRIIVEAD EKKGIVQASV TSSTHERKSK TYVITKNGKI 240
241 YLKHNSIAEE IPIAIVLKAC GILSDLEIMQ LVCGNDSSYQ DIFAVNLEES SKLDIYTQQQ 300
301 ALEYIGAKVK TMRRQKLTIL QEGIEAIATT VIAHLTVEAL DFREKALYIA MMTRRVVMAM 360
361 YNPKMIDDRD YVGNKRLELA GQLISLLFED LFKKFNNDFK LSIDKVLKKP NRAMEYDALL 420
421 SINVHSNNIT SGLNRAISTG NWSLKRFKME RAGVTHVLSR LSYISALGMM TRISSQFEKS 480
481 RKVSGPRALQ PSQFGMLCTA DTPEGEACGL VKNLALMTHI TTDDEEEPIK KLCYVLGVED 540
541 ITLIDSASLH LNYGVYLNGT LIGSIRFPTK FVTQFRHLRR TGKVSEFISI YSNSHQMAVH 600
601 IATDGGRICR PLIIVSDGQS RVKDIHLRKL LDGELDFDDF LKLGLVEYLD VNEENDSYIA 660
661 LYEKDIVPSM THLEIEPFTI LGAVAGLIPY PHHNQSPRNT YQCAMGKQAI GAIAYNQFKR 720
721 IDTLLYLMTY PQQPMVKTKT IELIDYDKLP AGQNATVAVM SYSGYDIEDA LVLNKSSIDR 780
781 GFGRCETRRK TTTVLKRYAN HTQDIIGGMR VDENGDPIWQ HQSLGPDGLG EVGMKVQSGQ 840
841 IYINKSVPTN SADAPNPNNV NVQTQYREAP VIYRGPEPSH IDQVMMSVSD NDQALIKVLL 900
901 RQNRRPELGD KFSSRHGQKG VCGIIVKQED MPFNDQGIVP DIIMNPHGFP SRMTVGKMIE 960
961 LISGKAGVLN GTLEYGTCFG GSKLEDMSKI LVDQGFNYSG KDMLYSGITG ECLQAYIFFG1020
1021 PIYYQKLKHM VLDKMHARAR GPRAVLTRQP TEGRSRDGGL RLGEMERDCV IAYGASQLLL1080
1081 ERLMISSDAF EVDVCDKCGL MGYSGWCTTC KSAENIIKMT IPYAAKLLFQ ELLSMNIAPR1140
1141 LRLEDIFQQ |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
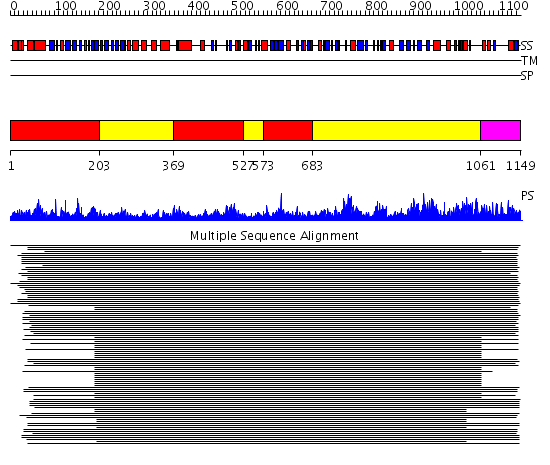
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..202] [369..526] [573..682] |
11000.0 | RBP2 | |
| 2 | View Details | [203..368] [527..572] [683..1060] |
11000.0 | RBP2 | |
| 3 | View Details | [1061..1149] | 11000.0 | RBP2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)