













| General Information: |
|
| Name(s) found: |
ACC1 /
YNR016C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2233 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum membrane
[IDA mitochondrion [IDA cytosol [TAS] |
| Biological Process: |
nuclear envelope organization
[TAS protein import into nucleus [IMP fatty acid biosynthetic process [TAS] |
| Molecular Function: |
acetyl-CoA carboxylase activity
[IMP biotin carboxylase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEESLFESS PQKMEYEITN YSERHTELPG HFIGLNTVDK LEESPLRDFV KSHGGHTVIS 60
61 KILIANNGIA AVKEIRSVRK WAYETFGDDR TVQFVAMATP EDLEANAEYI RMADQYIEVP 120
121 GGTNNNNYAN VDLIVDIAER ADVDAVWAGW GHASENPLLP EKLSQSKRKV IFIGPPGNAM 180
181 RSLGDKISST IVAQSAKVPC IPWSGTGVDT VHVDEKTGLV SVDDDIYQKG CCTSPEDGLQ 240
241 KAKRIGFPVM IKASEGGGGK GIRQVEREED FIALYHQAAN EIPGSPIFIM KLAGRARHLE 300
301 VQLLADQYGT NISLFGRDCS VQRRHQKIIE EAPVTIAKAE TFHEMEKAAV RLGKLVGYVS 360
361 AGTVEYLYSH DDGKFYFLEL NPRLQVEHPT TEMVSGVNLP AAQLQIAMGI PMHRISDIRT 420
421 LYGMNPHSAS EIDFEFKTQD ATKKQRRPIP KGHCTACRIT SEDPNDGFKP SGGTLHELNF 480
481 RSSSNVWGYF SVGNNGNIHS FSDSQFGHIF AFGENRQASR KHMVVALKEL SIRGDFRTTV 540
541 EYLIKLLETE DFEDNTITTG WLDDLITHKM TAEKPDPTLA VICGAATKAF LASEEARHKY 600
601 IESLQKGQVL SKDLLQTMFP VDFIHEGKRY KFTVAKSGND RYTLFINGSK CDIILRQLSD 660
661 GGLLIAIGGK SHTIYWKEEV AATRLSVDSM TTLLEVENDP TQLRTPSPGK LVKFLVENGE 720
721 HIIKGQPYAE IEVMKMQMPL VSQENGIVQL LKQPGSTIVA GDIMAIMTLD DPSKVKHALP 780
781 FEGMLPDFGS PVIEGTKPAY KFKSLVSTLE NILKGYDNQV IMNASLQQLI EVLRNPKLPY 840
841 SEWKLHISAL HSRLPAKLDE QMEELVARSL RRGAVFPARQ LSKLIDMAVK NPEYNPDKLL 900
901 GAVVEPLADI AHKYSNGLEA HEHSIFVHFL EEYYEVEKLF NGPNVREENI ILKLRDENPK 960
961 DLDKVALTVL SHSKVSAKNN LILAILKHYQ PLCKLSSKVS AIFSTPLQHI VELESKATAK1020
1021 VALQAREILI QGALPSVKER TEQIEHILKS SVVKVAYGSS NPKRSEPDLN ILKDLIDSNY1080
1081 VVFDVLLQFL THQDPVVTAA AAQVYIRRAY RAYTIGDIRV HEGVTVPIVE WKFQLPSAAF1140
1141 STFPTVKSKM GMNRAVSVSD LSYVANSQSS PLREGILMAV DHLDDVDEIL SQSLEVIPRH1200
1201 QSSSNGPAPD RSGSSASLSN VANVCVASTE GFESEEEILV RLREILDLNK QELINASIRR1260
1261 ITFMFGFKDG SYPKYYTFNG PNYNENETIR HIEPALAFQL ELGRLSNFNI KPIFTDNRNI1320
1321 HVYEAVSKTS PLDKRFFTRG IIRTGHIRDD ISIQEYLTSE ANRLMSDILD NLEVTDTSNS1380
1381 DLNHIFINFI AVFDISPEDV EAAFGGFLER FGKRLLRLRV SSAEIRIIIK DPQTGAPVPL1440
1441 RALINNVSGY VIKTEMYTEV KNAKGEWVFK SLGKPGSMHL RPIATPYPVK EWLQPKRYKA1500
1501 HLMGTTYVYD FPELFRQASS SQWKNFSADV KLTDDFFISN ELIEDENGEL TEVEREPGAN1560
1561 AIGMVAFKIT VKTPEYPRGR QFVVVANDIT FKIGSFGPQE DEFFNKVTEY ARKRGIPRIY1620
1621 LAANSGARIG MAEEIVPLFQ VAWNDAANPD KGFQYLYLTS EGMETLKKFD KENSVLTERT1680
1681 VINGEERFVI KTIIGSEDGL GVECLRGSGL IAGATSRAYH DIFTITLVTC RSVGIGAYLV1740
1741 RLGQRAIQVE GQPIILTGAP AINKMLGREV YTSNLQLGGT QIMYNNGVSH LTAVDDLAGV1800
1801 EKIVEWMSYV PAKRNMPVPI LETKDTWDRP VDFTPTNDET YDVRWMIEGR ETESGFEYGL1860
1861 FDKGSFFETL SGWAKGVVVG RARLGGIPLG VIGVETRTVE NLIPADPANP NSAETLIQEP1920
1921 GQVWHPNSAF KTAQAINDFN NGEQLPMMIL ANWRGFSGGQ RDMFNEVLKY GSFIVDALVD1980
1981 YKQPIIIYIP PTGELRGGSW VVVDPTINAD QMEMYADVNA RAGVLEPQGM VGIKFRREKL2040
2041 LDTMNRLDDK YRELRSQLSN KSLAPEVHQQ ISKQLADRER ELLPIYGQIS LQFADLHDRS2100
2101 SRMVAKGVIS KELEWTEARR FFFWRLRRRL NEEYLIKRLS HQVGEASRLE KIARIRSWYP2160
2161 ASVDHEDDRQ VATWIEENYK TLDDKLKGLK LESFAQDLAK KIRSDHDNAI DGLSEVIKML2220
2221 STDDKEKLLK TLK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ACC1 HFA1 |
| View Details | Gavin AC, et al. (2002) |
|
ACC1 AVT4 BEM2 CCT2 CCT5 CCT8 CDC25 CLU1 CPA2 FAB1 FDH1 GCN20 GFA1 GUS1 HRK1 ILV1 KAP123 LYS12 MDS3 MIA40 MRPL3 PSA1 RPA135 RPB2 RPN10 RPN11 RPN12 RPN8 RPT3 RPT5 RPT6 SAM1 SAP155 SAP185 SAP190 SEC27 SIT4 SMC3 SWT21 TCB1 TIF1, TIF2 TSA1 TUB3 URA7 VAC14 VAS1 YOL098C |
| View Details | Ho Y, et al. (2002) |

|
ACC1 ACH1 ADH2 ADR1 AHA1 ARC35 ARO4 ARP2 ATP3 BGL2 CCT3 CCT6 CDC60 COP1 CPA2 CPR6 DEF1 DMC1 DPM1 FAA1 FAA4 FOL2 GAL7 GCD11 GFA1 GLT1 GND1 GSF2 GUS1 IDH1 IDH2 ILS1 ISA2 KGD2 LIA1 LSC1 LST8 LYS12 MAE1 MDH1 MDJ1 MET18 NDE1 NGL2 NOP14 NPA3 OLA1 PHO86 POR1 PYC1 PYC2 RET1 RMT2 RNR2 RPA135 RPA190 RPA49 RPB5 RPC19 RPC25 RPC34 RPC40 RPC82 RPN12 RPN3 RPO26 RPO31 RPT4 RRP3 RVB2 RVS167 SAP155 SAP185 SAP190 SEC27 SEC53 SGD1 SHS1 SIP1 SIT4 SLC1 SMC4 STI1 TAP42 TBS1 TFP1 TRP3 TRP5 TSA2 URE2 UTP5 VMA4 YBT1 YER138C YFL042C YHR033W YHR112C YKU80 YML020W YMR196W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Boi 2 gst control | McCusker D, et al (2007) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
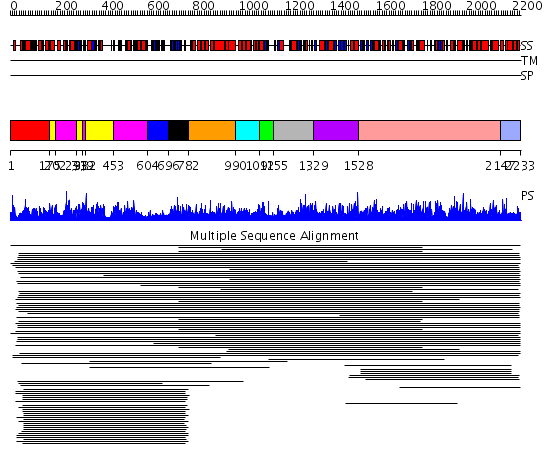
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | 700.228787 | Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), C-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase, (BC), N-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), domain 2 | |
| 2 | View Details | [175..201] [293..318] [332..452] |
700.228787 | Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), C-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase, (BC), N-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), domain 2 | |
| 3 | View Details | [202..292] [319..331] [453..603] |
700.228787 | Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), C-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase, (BC), N-domain; Biotin carboxylase subunit of acetyl-CoA carboxylase (BC), domain 2 | |
| 4 | View Details | [604..695] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [696..781] | 12.522879 | Biotin carboxyl carrier domain of transcarboxylase (TC 1.3S) | |
| 6 | View Details | [782..989] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [990..1091] | 1.037977 | View MSA. Confident ab initio structure predictions are available. | |
| 8 | View Details | [1092..1154] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1155..1328] | 1.036981 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1329..1527] | 1.04297 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1528..2146] | 56.143897 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [2147..2233] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)