













| General Information: |
|
| Name(s) found: |
MRPL3 /
YMR024W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 390 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial large ribosomal subunit [IPI |
| Biological Process: |
translation
[TAS |
| Molecular Function: |
structural constituent of ribosome
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIVLKRAIA AGMKPFPNST WHWSRTIRPF SQHLSSTCFL QQSSRFTSKR YLHLSTLTQQ 60
61 EKRFLPESEL AKYKEYYQGL KSTVNEIPES VASKSPSLRT LHKRLQLPNE LTYSTLSRCL 120
121 TCPSAKLPDK INNPTKGAAF VNTVPTNKYL DNHGLNIMGK NLLSYHVTKS IIQKYPRLPT 180
181 VVLNAAVNAY ISEAVLAHIA KYWGIEVETT SVLSRYLKME PFEFTLGRLK FFNNSLNSKD 240
241 GIELITGKNF SETSALAMSV RSIIAAIWAV TEQKDSQAVY RFIDDHIMSR KLDITKMFQF 300
301 EQPTRELAML CRREGLEKPV SKLVAESGRL SKSPVFIVHV FSGEETLGEG YGSSLKEAKA 360
361 RAATDALMKW YCYEPLAQQE PVIDPGTVVV |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #33 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #35 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
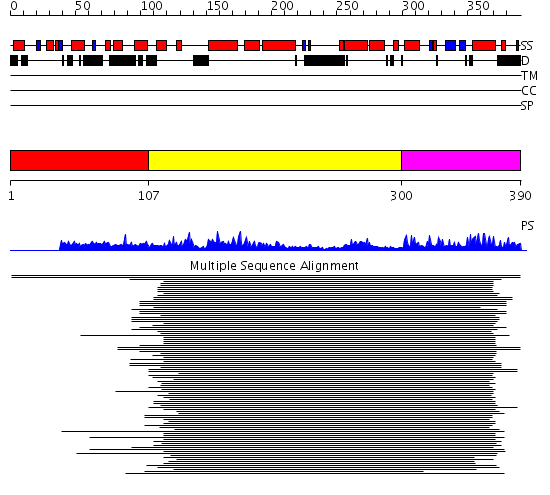
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 1.037993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [107..299] | 22.221849 | RNase III endonuclease catalytic domain | |
| 3 | View Details | [300..390] | 12.39794 | Double-stranded RNA-binding protein A, second dsRBD |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)