













| General Information: |
|
| Name(s) found: |
CWC22 /
YGR278W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 577 amino acids |
Gene Ontology: |
|
| Cellular Component: |
spliceosomal complex
[IPI |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IPI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTATIQDED IKFQRENWEM IRSHVSPIIS NLTMDNLQES HRDLFQVNIL IGRNIICKNV 60
61 VDFTLNKQNG RLIPALSALI ALLNSDIPDI GETLAKELML MFVQQFNRKD YVSCGNILQC 120
121 LSILFLYDVI HEIVILQILL LLLEKNSLRL VIAVMKICGW KLALVSKKTH DMIWEKLRYI 180
181 LQTQELSSTL RESLETLFEI RQKDYKSGSQ GLFILDPTSY TVHTHSYIVS DEDEANKELG 240
241 NFEKCENFNE LTMAFDTLRQ KLLINNTSDT NEGSNSQLQI YDMTSTNDVE FKKKIYLVLK 300
301 SSLSGDEAAH KLLKLKIANN LKKSVVDIII KSSLQESTFS KFYSILSERM ITFHRSWQTA 360
361 YNETFEQNYT QDIEDYETDQ LRILGKFWGH LISYEFLPMD CLKIIKLTEE ESCPQGRIFI 420
421 KFLFQELVNE LGLDELQLRL NSSKLDGMFP LEGDAEHIRY SINFFTAIGL GLLTEDMRSR 480
481 LTIIQEVEDA EEEEKKLREE EELEKLRKKA RESQPTQGPK IHESRLFLQK DTRENSRSRS 540
541 PFTVETRKRA RSRTPPRGSR NHRNRSRTPP ARRQRHR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Unpublished Data |
| View Data |
|
Huh WK, et al. (2003) |
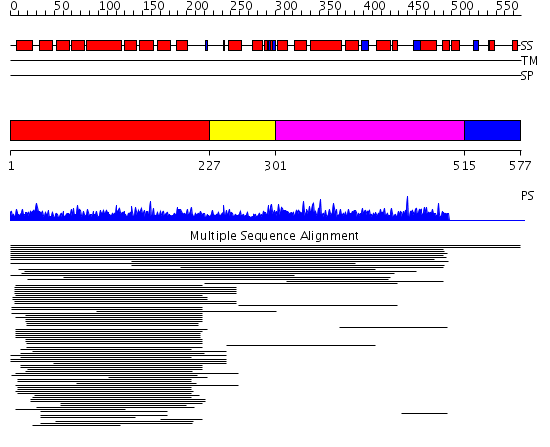
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..226] | 45.0 | Eukaryotic initiation factor eIF4G | |
| 2 | View Details | [227..300] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [301..514] | 7.46 | CBP80, 80KDa nuclear cap-binding protein | |
| 4 | View Details | [515..577] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)