













| General Information: |
|
| Name(s) found: |
PRP45 /
YAL032C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 379 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spliceosomal complex [IPI |
| Biological Process: |
positive regulation of transcription from RNA polymerase II promoter
[IDA nuclear mRNA splicing, via spliceosome [IMP |
| Molecular Function: |
transcription activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSNRLPPPK HSQGRVSTAL SSDRVEPAIL TDQIAKNVKL DDFIPKRQSN FELSVPLPTK 60
61 AEIQECTART KSYIQRLVNA KLANSNNRAS SRYVTETHQA PANLLLNNSH HIEVVSKQMD 120
121 PLLPRFVGKK ARKVVAPTEN DEVVPVLHMD GSNDRGEADP NEWKIPAAVS NWKNPNGYTV 180
181 ALERRVGKAL DNENNTINDG FMKLSEALEN ADKKARQEIR SKMELKRLAM EQEMLAKESK 240
241 LKELSQRARY HNGTPQTGAI VKPKKQTSTV ARLKELAYSQ GRDVSEKIIL GAAKRSEQPD 300
301 LQYDSRFFTR GANASAKRHE DQVYDNPLFV QQDIESIYKT NYEKLDEAVN VKSEGASGSH 360
361 GPIQFTKAES DDKSDNYGA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
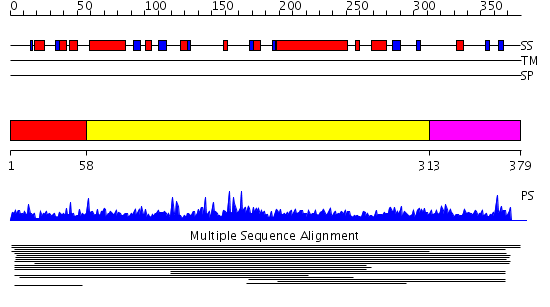
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..57] | 1.013991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [58..312] | 109.853872 | SKIP/SNW domain No confident structure predictions are available. | |
| 3 | View Details | [313..379] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [298..379] | 4.029995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
|||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)