













| General Information: |
|
| Name(s) found: |
SYF1 /
YDR416W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: |
spliceosomal complex
[IDA |
| Biological Process: |
cell cycle
[TAS nuclear mRNA splicing, via spliceosome [IPI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAYIAMKGV ITNVDENIRN DEDVAFEYEI QKTPQNILTW KRYIEYWKEE GRTDKQIRWL 60
61 YERFCSQFVT DTSIWEDYIR WESTKEVVET SRIFWLFQRC LKSCVRDCDR ICLSYLELAI 120
121 EQYDLAMIRH ALASSLMKME REMHRKVWDP VIKFVEEKVL PLTQLDSTQE DEEESTDEAE 180
181 LINVLLVKGF TKGGFISEEI SENGSRGDIW SSHILERYLK VAPQQKRNES LATLALTRDN 240
241 ITIKSVYEKY LPQDENSGKY LPSSELPFEL NFNYLASLEK LGLDNQYEEF MRQMNGIYPD 300
301 KWLFLILSLA KYYISRGRLD SCGDLLKKSL QQTLRYSDFD RIYNFYLLFE QECSQFILGK 360
361 LKENDSKFFN QKDWTEKLQA HMATFESLIN LYDIYLNDVA LRQDSNLVET WMKRVSLQKS 420
421 AAEKCNVYSE AILKIDPRKV GTPGSFGRLW CSYGDLYWRS NAISTARELW TQSLKVPYPY 480
481 IEDLEEIYLN WADRELDKEG VERAFSILED ALHVPTNPEI LLEKYKNGHR KIPAQTVLFN 540
541 SLRIWSKYID YLEAYCPKDA NSSDKIFNKT KMAYNTVIDL RLITPAMAEN FALFLQNHYE 600
601 VMESFQVYEK TIPLFPPEIQ YELWIEYLEV ATSHQLSSLS PEHIRFLFEK ALKNLCSNGI 660
661 DCKTIFIAYS VFEERISGLI SKSIEILRRG AVIGTVSVST HLESRLQLWR MCISKAESTL 720
721 GPSVTRELYQ ECIQILPNSK AVEFVIKFSD FESSIGETIR AREILAYGAK LLPPSRNTEL 780
781 WDSFEIFELK HGDKETYKDM LKMKKVLESN MLIDSASVSH EEGNINFVAA ATSHAPNSHT 840
841 LTQSTSSYSI NPDEIELDI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
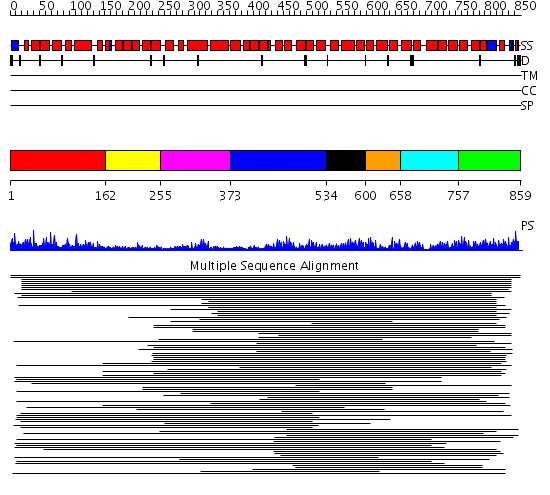
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 14.67 | Protein farnesyltransferase alpha-subunit | |
| 2 | View Details | [162..254] | 14.67 | Protein farnesyltransferase alpha-subunit | |
| 3 | View Details | [255..372] | 14.67 | Protein farnesyltransferase alpha-subunit | |
| 4 | View Details | [373..533] | 16.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 5 | View Details | [534..599] | 16.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 6 | View Details | [600..657] | 16.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 7 | View Details | [658..756] | 10.95 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 8 | View Details | [757..859] | 10.95 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)