













| General Information: |
|
| Name(s) found: |
URE2 /
YNL229C
[SGD]
|
| Description(s) found:
Found 55 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 354 amino acids |
Gene Ontology: |
|
| Cellular Component: |
soluble fraction
[IDA cytosol [IDA |
| Biological Process: |
response to aluminum ion
[IMP telomere maintenance [IMP regulation of nitrogen utilization [IGI protein urmylation [IMP cytoplasmic sequestering of transcription factor [IMP |
| Molecular Function: |
transcription corepressor activity
[IGI phosphoprotein binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMNNNGNQVS NLSNALRQVN IGNRNSNTTT DQSNINFEFS TGVNNNNNNN SSSNNNNVQN 60
61 NNSGRNGSQN NDNENNIKNT LEQHRQQQQA FSDMSHVEYS RITKFFQEQP LEGYTLFSHR 120
121 SAPNGFKVAI VLSELGFHYN TIFLDFNLGE HRAPEFVSVN PNARVPALID HGMDNLSIWE 180
181 SGAILLHLVN KYYKETGNPL LWSDDLADQS QINAWLFFQT SGHAPMIGQA LHFRYFHSQK 240
241 IASAVERYTD EVRRVYGVVE MALAERREAL VMELDTENAA AYSAGTTPMS QSRFFDYPVW 300
301 LVGDKLTIAD LAFVPWNNVV DRIGINIKIE FPEVYKWTKH MMRRPAVIKA LRGE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
GLN3 MET4 URE2 |
| View Details | Ho Y, et al. (2002) |

|
ACC1 AHA1 ARO4 CDC28 CDC60 CEG1 CKA1 CKA2 CKB1 CKB2 FKH1 FYV8 GCD2 GCD7 GCN3 HHF1, HHF2 HTB1 ILS1 MBP1 MGM101 MPH1 NET1 NOP1 RPA135 RPA190 RPC40 RRP1 SEC2 SIN3 SMC4 SUI2 SUI3 TRP5 UBP12 URE2 VMA4 YGR017W YMR144W |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
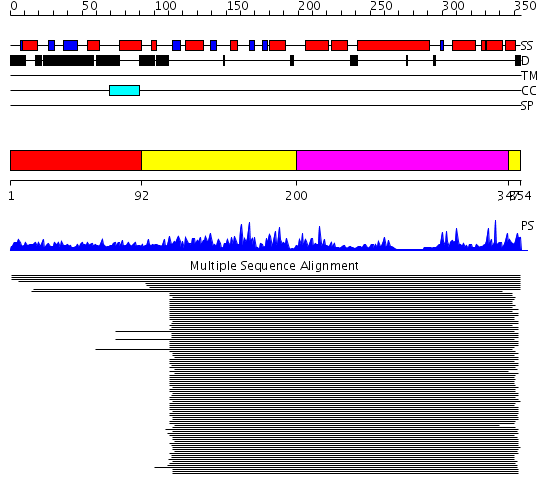
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | 1.066984 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [92..199] [347..354] |
1625.39794 | Yeast prion protein ure2p, nitrogen regulation fragment | |
| 3 | View Details | [200..346] | 1625.39794 | Yeast prion protein ure2p, nitrogen regulation fragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)