













| General Information: |
|
| Name(s) found: |
TRM82 /
YDR165W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 444 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
ribosome biogenesis
[RCA tRNA methylation [IMP |
| Molecular Function: |
protein binding
[IPI tRNA (guanine-N7-)-methyltransferase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVIHPLQNL LTSRDGSLVF AIIKNCILSF KYQSPNHWEF AGKWSDDFDK IQESRNTTAK 60
61 EQQGQSSENE NENKKLKSNK GDSIKRTAAK VPSPGLGAPP IYSYIRNLRL TSDESRLIAC 120
121 ADSDKSLLVF DVDKTSKNVL KLRKRFCFSK RPNAISIAED DTTVIIADKF GDVYSIDINS 180
181 IPEEKFTQEP ILGHVSMLTD VHLIKDSDGH QFIITSDRDE HIKISHYPQC FIVDKWLFGH 240
241 KHFVSSICCG KDYLLLSAGG DDKIFAWDWK TGKNLSTFDY NSLIKPYLND QHLAPPRFQN 300
301 ENNDIIEFAV SKIIKSKNLP FVAFFVEATK CIIILEMSEK QKGDLALKQI ITFPYNVISL 360
361 SAHNDEFQVT LDNKESSGVQ KNFAKFIEYN LNENSFVVNN EKSNEFDSAI IQSVQGDSNL 420
421 VTKKEEIYPL YNVSSLRKHG EHYS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
TRM8 TRM82 |
| View Details | Krogan NJ, et al. (2006) |
|
BNA5 HXK1 TRM8 TRM82 |
| View Details | Qiu et al. (2008) |
|
GCD10 GCD14 TRM82 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
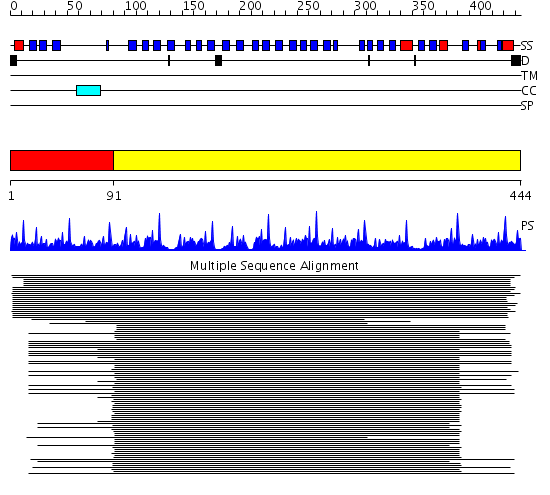
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 58.522879 | Tup1, C-terminal domain | |
| 2 | View Details | [91..444] | 77.09691 | beta1-subunit of the signal-transducing G protein heterotrimer |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)