













| General Information: |
|
| Name(s) found: |
CEG1 /
YGL130W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 459 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI |
| Biological Process: |
mRNA capping
[TAS positive regulation of transcription from RNA polymerase II promoter [IGI |
| Molecular Function: |
mRNA guanylyltransferase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLAMESRVA PEIPGLIQPG NVTQDLKMMV CKLLNSPKPT KTFPGSQPVS FQHSDVEEKL 60
61 LAHDYYVCEK TDGLRVLMFI VINPVTGEQG CFMIDRENNY YLVNGFRFPR LPQKKKEELL 120
121 ETLQDGTLLD GELVIQTNPM TKLQELRYLM FDCLAINGRC LTQSPTSSRL AHLGKEFFKP 180
181 YFDLRAAYPN RCTTFPFKIS MKHMDFSYQL VKVAKSLDKL PHLSDGLIFT PVKAPYTAGG 240
241 KDSLLLKWKP EQENTVDFKL ILDIPMVEDP SLPKDDRNRW YYNYDVKPVF SLYVWQGGAD 300
301 VNSRLKHFDQ PFDRKEFEIL ERTYRKFAEL SVSDEEWQNL KNLEQPLNGR IVECAKNQET 360
361 GAWEMLRFRD DKLNGNHTSV VQKVLESIND SVSLEDLEEI VGDIKRCWDE RRANMAGGSG 420
421 RPLPSQSQNA TLSTSKPVHS QPPSNDKEPK YVDEDDWSD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CEG1 CET1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
CEG1 CET1 |
| View Details | Krogan NJ, et al. (2006) |
|
CEG1 CET1 YHR130C |
| View Details | Gavin AC, et al. (2006) |

|
CEG1 CET1 |
| View Details | Gavin AC, et al. (2002) |
|
AVO1 BEM2 CEG1 CET1 CLU1 CTR9 HHF1, HHF2 LSM12 NIP1 NOP1 PDB1 PRP43 RPG1 SPT16 SSA3 SSA4 TOP2 VPS1 |
| View Details | Ho Y, et al. (2002) |

|
CDC28 CEG1 CKA1 CKA2 CKB1 CKB2 FKH1 FYV8 GCD2 GCD7 GCN3 HHF1, HHF2 HTB1 MBP1 MGM101 MPH1 NET1 NOP1 RRP1 SEC2 SIN3 SUI2 SUI3 UBP12 URE2 YGR017W YMR144W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
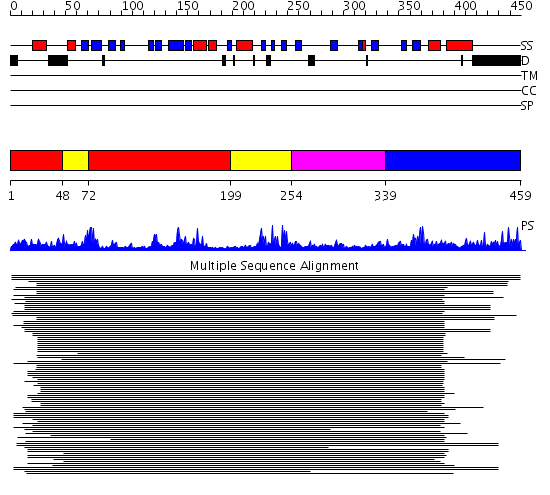
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..47] [72..198] |
149.218487 | RNA guanylyltransferase (mRNA capping enzyme); RNA guanylyltransferase (mRNA capping enzyme), N-terminal domain | |
| 2 | View Details | [48..71] [199..253] |
149.218487 | RNA guanylyltransferase (mRNA capping enzyme); RNA guanylyltransferase (mRNA capping enzyme), N-terminal domain | |
| 3 | View Details | [254..338] | 149.218487 | RNA guanylyltransferase (mRNA capping enzyme); RNA guanylyltransferase (mRNA capping enzyme), N-terminal domain | |
| 4 | View Details | [339..459] | 10.46 | RNA guanylyltransferase (mRNA capping enzyme); RNA guanylyltransferase (mRNA capping enzyme), N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)