













| General Information: |
|
| Name(s) found: |
TRP3 /
YKL211C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 484 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA anthranilate synthase complex [IPI |
| Biological Process: |
tryptophan biosynthetic process
[TAS |
| Molecular Function: |
indole-3-glycerol-phosphate synthase activity
[IDA anthranilate synthase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVHAATNPI NKHVVLIDNY DSFTWNVYEY LCQEGAKVSV YRNDAITVPE IAALNPDTLL 60
61 ISPGPGHPKT DSGISRDCIR YFTGKIPVFG ICMGQQCMFD VFGGEVAYAG EIVHGKTSPI 120
121 SHDNCGIFKN VPQGIAVTRY HSLAGTESSL PSCLKVTAST ENGIIMGVRH KKYTVEGVQF 180
181 HPESILTEEG HLMIRNILNV SGGTWEENKS SPSNSILDRI YARRKIDVNE QSKIPGFTFQ 240
241 DLQSNYDLGL APPLQDFYTV LSSSHKRAVV LAEVKRASPS KGPICLKAVA AEQALKYAEA 300
301 GASAISVLTE PHWFHGSLQD LVNVRKILDL KFPPKERPCV LRKEFIFSKY QILEARLAGA 360
361 DTVLLIVKML SQPLLKELYS YSKDLNMEPL VEVNSKEELQ RALEIGAKVV GVNNRDLHSF 420
421 NVDLNTTSNL VESIPKDVLL IALSGITTRD DAEKYKKEGV HGFLVGEALM KSTDVKKFIH 480
481 ELCE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
TRP2 TRP3 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
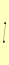
|
TRP2 TRP3 |
| View Details | Krogan NJ, et al. (2006) |

|
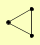
STB3 TRP2 TRP3 |
| View Details | Gavin AC, et al. (2006) |
|
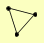
SAM1 TRP2 TRP3 |
| View Details | Gavin AC, et al. (2002) |
|
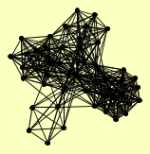
SAM1 TRP2 TRP3 |
| View Details | Ho Y, et al. (2002) |
|
ACC1 ADR1 ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 ATP3 BGL2 BNI1 CBP6 COP1 DMC1 DPM1 DUR1,2 FRS2 GCD11 GCN3 HXT7 IMH1 KAP122 MCM4 MCM7 MCX1 MDJ1 MET18 NDE1 NOP14 NPA3 PET10 PET9 PHO86 PRB1 RET2 RPT4 RVB2 SEC21 SEC26 SEC27 SEC28 SLC1 TRP3 TRP5 VPS8 YJR029W |
| View Details | Qiu et al. (2008) |

|
TRP2 TRP3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
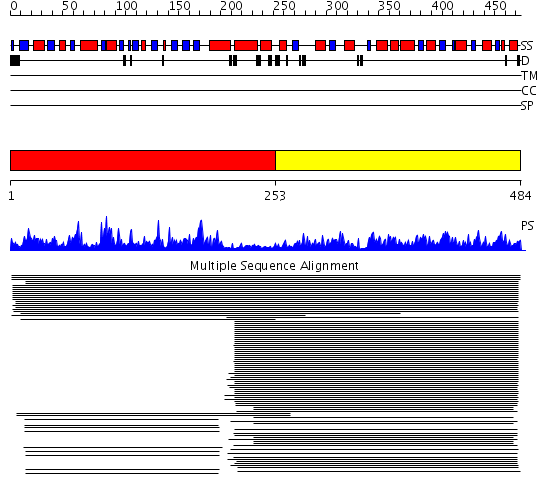
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..252] | 299.454997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [253..484] | 387.201249 | Indole-3-glycerophosphate synthase, IPGS |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)