













| General Information: |
|
| Name(s) found: |
RPB7 /
YDR404C
[SGD]
|
| Description(s) found:
Found 68 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 171 amino acids |
Gene Ontology: |
|
| Cellular Component: |
DNA-directed RNA polymerase II, core complex
[TAS |
| Biological Process: |
transcription from RNA polymerase II promoter
[TAS |
| Molecular Function: |
DNA-directed RNA polymerase activity
[TAS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
DST1 IWR1 RPB10 RPB11 RPB2 RPB3 RPB4 RPB5 RPB7 RPB8 RPB9 RPC10 RPO21 RPO26 SPT4 SPT5 TAF14 TFG1 TFG2 |
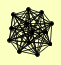
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CDC73 RPB10 RPB11 RPB2 RPB3 RPB4 RPB5 RPB7 RPB8 RPB9 RPC10 RPO21 RPO26 |
| View Details | Krogan NJ, et al. (2006) |

|
ALG3 BUD27 CBF2 DST1 ESS1 IWR1 RPB10 RPB11 RPB2 RPB3 RPB4 RPB5 RPB7 RPB8 RPB9 RPO21 RPO26 SPT4 TAF14 TFG1 TFG2 YBL059W YBR259W YPL225W |
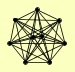
| View Details | Gavin AC, et al. (2006) |
|
IWR1 RPB11 RPB2 RPB3 RPB4 RPB7 RPB9 TFG1 TFG2 |
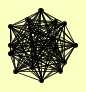
| View Details | Gavin AC, et al. (2002) |
|
ABD1 AOS1 IWR1 RPB11 RPB2 RPB3 RPB4 RPB5 RPB7 RPB8 RPB9 RPO21 RPO26 SPT4 SPT5 TAF14 TFG1 TFG2 |
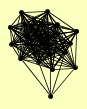
| View Details | Qiu et al. (2008) |
|
RET1 RPA12 RPA135 RPA190 RPA43 RPB10 RPB11 RPB2 RPB3 RPB4 RPB5 RPB7 RPB8 RPB9 RPC10 RPC17 RPC31 RPC37 RPC40 RPO21 RPO26 RPO31 TAF14 TFB5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | 116.228787 | C-terminal domain of RNA polymerase II subunit RBP4 (RpoE); N-terminal, heterodimerisation domain of RBP4 (RpoE) | |
| 2 | View Details | [80..171] | 116.228787 | C-terminal domain of RNA polymerase II subunit RBP4 (RpoE); N-terminal, heterodimerisation domain of RBP4 (RpoE) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)