













| General Information: |
|
| Name(s) found: |
HRP1 /
YOL123W
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 534 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA mRNA cleavage factor complex [IPI |
| Biological Process: |
mRNA polyadenylation
[IDA mRNA cleavage [IDA |
| Molecular Function: |
RNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSDEEDFND IYGDDKPTTT EEVKKEEEQN KAGSGTSQLD QLAALQALSS SLNKLNNPNS 60
61 NNSSSNNSNQ DTSSSKQDGT ANDKEGSNED TKNEKKQESA TSANANANAS SAGPSGLPWE 120
121 QLQQTMSQFQ QPSSQSPPQQ QVTQTKEERS KADLSKESCK MFIGGLNWDT TEDNLREYFG 180
181 KYGTVTDLKI MKDPATGRSR GFGFLSFEKP SSVDEVVKTQ HILDGKVIDP KRAIPRDEQD 240
241 KTGKIFVGGI GPDVRPKEFE EFFSQWGTII DAQLMLDKDT GQSRGFGFVT YDSADAVDRV 300
301 CQNKFIDFKD RKIEIKRAEP RHMQQKSSNN GGNNGGNNMN RRGGNFGNQG DFNQMYQNPM 360
361 MGGYNPMMNP QAMTDYYQKM QEYYQQMQKQ TGMDYTQMYQ QQMQQMAMMM PGFAMPPNAM 420
421 TLNQPQQDSN ATQGSPAPSD SDNNKSNDVQ TIGNTSNTDS GSPPLNLPNG PKGPSQYNDD 480
481 HNSGYGYNRD RGDRDRNDRD RDYNHRSGGN HRRNGRGGRG GYNRRNNGYH PYNR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
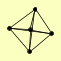
|
ENP1 HRP1 KAP104 NAB2 YNL035C |
| View Details | (MIPS) Mewes HW, et al. (2004) |
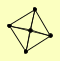
|
CLP1 HRP1 PCF11 RNA14 RNA15 |
| View Details | Gavin AC, et al. (2006) |
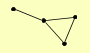
|
AEP1 HRP1 KAP104 RPS9A |
| View Details | Gavin AC, et al. (2002) |
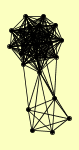
|
AEP1 AEP2 ATP22 BMH1 BMH2 CLU1 CRM1 ECM29 FKS1 GCN1 GEA2 GFA1 HRP1 KAP104 KAP123 LCB1 LCB2 LYS12 MIR1 NAB2 SAM1 SAM2 SES1 TSR1 YHR020W |
| View Details | Ho Y, et al. (2002) |

|
ADH4 ADR1 ARC1 AYR1 BGL2 CBP6 DIM1 DPS1 ENP1 FAA1 GAR1 GSP1 GSP2 HEM15 HRP1 HTA2 KAP104 KTR3 MRT4 NAB2 NDE1 OAC1 PAA1 PET10 PET9 PGM2 PSD1 RPT4 RTN2 SAC1 TFG2 VMA8 YDL063C YHM2 YNL035C YRB1 |
| View Details | Qiu et al. (2008) |
|
CFT1 CFT2 CLP1 FIP1 HRP1 PAP1 PFS2 PTA1 PTI1 REF2 RNA14 RNA15 SSU72 SWD2 YSH1 YTH1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
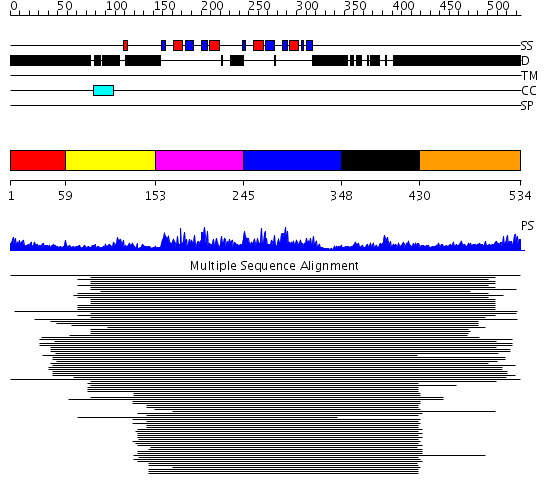
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..58] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [59..152] | 167.39794 | Poly(A)-binding protein | |
| 3 | View Details | [153..244] | 402.9794 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 4 | View Details | [245..347] | 13.0 | Polypyrimidine tract-binding protein | |
| 5 | View Details | [348..429] | 13.0 | Polypyrimidine tract-binding protein | |
| 6 | View Details | [430..534] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)