













| General Information: |
|
| Name(s) found: |
DIM1 /
YPL266W
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 318 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear outer membrane
[TAS nucleolus [TAS |
| Biological Process: |
rRNA modification
[TAS ribosome biogenesis [RCA |
| Molecular Function: |
rRNA (adenine-N6,N6-)-dimethyltransferase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKAAKKKYS GATSSKQVSA EKHLSSVFKF NTDLGQHILK NPLVAQGIVD KAQIRPSDVV 60
61 LEVGPGTGNL TVRILEQAKN VVAVEMDPRM AAELTKRVRG TPVEKKLEIM LGDFMKTELP 120
121 YFDICISNTP YQISSPLVFK LINQPRPPRV SILMFQREFA LRLLARPGDS LYCRLSANVQ 180
181 MWANVTHIMK VGKNNFRPPP QVESSVVRLE IKNPRPQVDY NEWDGLLRIV FVRKNRTISA 240
241 GFKSTTVMDI LEKNYKTFLA MNNEMVDDTK GSMHDVVKEK IDTVLKETDL GDKRAGKCDQ 300
301 NDFLRLLYAF HQVGIHFS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
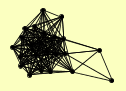
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
DIM1 DIP2 EMG1 ENP1 HRR25 IMP3 LTV1 MPP10 NAN1 NOC4 PNO1 PWP2 RIO2 RPS13 RPS9A RPS9B RRP12 TSR1 UTP10 UTP4 UTP6 UTP7 UTP8 |
| View Details | Gavin AC, et al. (2002) |

|
ADE4 ADH1 ARP9 ASC1 BEM2 BFR2 BMS1 BRX1 BUD21 CBF5 CKA1 CKA2 CKB1 CKB2 CMS1 DBP8 DIA4 DIM1 DIP2 DIS3 ECM16 ECM29 EMG1 ENP1 ENP2 ERO1 ESF1 ESF2 FAP7 GCN1 HCA4 HHF1, HHF2 HRR25 HSL1 HTA2 IMP3 IMP4 KRE33 KRR1 LCP5 LTV1 MPP10 MRD1 MVD1 NAN1 NOB1 NOC2 NOC4 NOP1 NOP14 NOP56 NOP58 NOP6 PNO1 PRE6 PRE9 PRP43 PWP2 RCL1 RIO2 RML2 ROK1 RPP2B RRP12 RRP7 RRP9 RTG2 SAM1 SCL1 SEC21 SLX9 SNF12 SNF2 SNF5 SNF6 SOF1 SWI1 SWI3 SWP82 TIF1, TIF2 TSR1 UTP10 UTP11 UTP13 UTP15 UTP18 UTP20 UTP21 UTP22 UTP30 UTP4 UTP6 UTP7 UTP8 UTP9 VPS1 YGR054W YOR059C |
| View Details | Ho Y, et al. (2002) |
|
DIM1 ENP1 HRP1 KAP104 MRT4 NAB2 YNL035C |
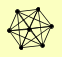
| View Details | Qiu et al. (2008) |

|
DIM1 NOP1 TRM112 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | 115.9897 | rRNA adenine dimethylase | |
| 2 | View Details | [216..318] | 115.9897 | rRNA adenine dimethylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)