













| General Information: |
|
| Name(s) found: |
SNF12 /
YNR023W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 566 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SWI/SNF complex
[TAS chromatin remodeling complex [TAS |
| Biological Process: |
chromatin remodeling
[TAS |
| Molecular Function: |
general RNA polymerase II transcription factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKVMKPSNG KGSRKSSKAA TPDTKNFFHA KKKDPVNQDK ANNASQITPT VPHSHPSDMV 60
61 IPDHLAELIP ELYSFQQLVD SEKRLDHFIH LRNLHMKRMV AQWERSKLSQ EFLYPHLNFP 120
121 NVKFLRIFIS NVSENQPWQM DTNNEADLMA LENATWTMRI EGRLLDNVQA NDPAREKFSS 180
181 FIESIVVDFK NKENDNVPST KFNAAPEENA TEGPSDKKLN LNLPLQFSLP NGDNSTTTNT 240
241 DQNNATMGEE TAKKDMSSTT PKLESVKWQY DPNNPVDFDG LDIKRVGSEN VECTISILRK 300
301 SSPEEPFMSY SPQLTAIIGL KSGTSHDAIF SIYKYIHLNE LLTNDESAFE NLMGNRNNHN 360
361 SNTSTSKMLD AASSQVSIVK LDTQLITLLP SSLKESSPDT MKLTDLLSLI NSTHLLPLQP 420
421 IEIDYTVRVD KASTYGELVL DIEVPDVNAL KFNNTQRESQ IGAAELNENA RELEQIKPKI 480
481 ALQDKEITSV LSNLHESNKR YRFFKKISED PVKALNECIA STSNALKVLS GDEGYNEDMV 540
541 RRANFYKENE AMLRENIEVI LSNGRM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
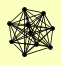
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP7 ARP9 SNF11 SNF12 SNF2 SNF5 SNF6 SWI1 SWI3 SWP82 |
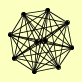
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ARP7 ARP9 SNF11 SNF12 SNF2 SNF5 SNF6 SWI1 SWI3 TAF14 |
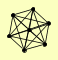
| View Details | Krogan NJ, et al. (2006) |
|
SNF12 SNF2 SNF5 SNF6 SWI1 SWI3 SWP82 |
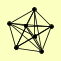
| View Details | Gavin AC, et al. (2006) |
|
ARP9 SNF11 SNF12 SNF5 SNF6 SWI1 SWI3 |
| View Details | Gavin AC, et al. (2002) |
|
ARP9 BEM2 DIM1 ENP1 HRR25 LTV1 NOB1 PNO1 RIO2 RRP12 SNF12 SNF2 SNF5 SNF6 SWI1 SWI3 SWP82 TSR1 VPS1 YGR054W |
| View Details | Qiu et al. (2008) |

|
ARP7 ARP9 SNF11 SNF12 SNF2 SNF5 SNF6 SWI1 SWI3 SWP82 TAF14 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
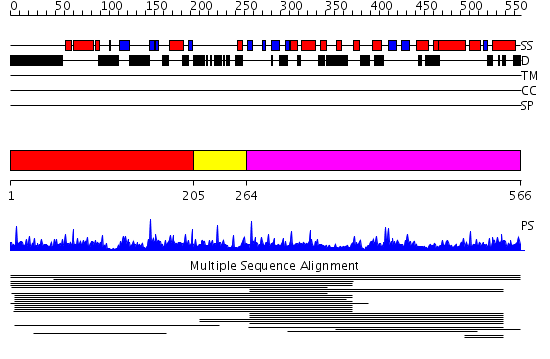
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..204] | 1.012974 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [205..263] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [264..566] | 5.016935 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [305..450] | 22.0 | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | |
| 5 | View Details | [451..566] | 1.039978 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)