













| General Information: |
|
| Name(s) found: |
ARP7 /
YPR034W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 477 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA SWI/SNF complex [IPI RSC complex [IDA |
| Biological Process: |
ATP-dependent chromatin remodeling
[IDA |
| Molecular Function: |
DNA-dependent ATPase activity
[IDA general RNA polymerase II transcription factor activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLNRKCVVI HNGSHRTVAG FSNVELPQCI IPSSYIKRTD EGGEAEFIFG TYNMIDAAAE 60
61 KRNGDEVYTL VDSQGLPYNW DALEMQWRYL YDTQLKVSPE ELPLVITMPA TNGKPDMAIL 120
121 ERYYELAFDK LNVPVFQIVI EPLAIALSMG KSSAFVIDIG ASGCNVTPII DGIVVKNAVV 180
181 RSKFGGDFLD FQVHERLAPL IKEENDMENM ADEQKRSTDV WYEASTWIQQ FKSTMLQVSE 240
241 KDLFELERYY KEQADIYAKQ QEQLKQMDQQ LQYTALTGSP NNPLVQKKNF LFKPLNKTLT 300
301 LDLKECYQFA EYLFKPQLIS DKFSPEDGLG PLMAKSVKKA GASINSMKAN TSTNPNGLGT 360
361 SHINTNVGDN NSTASSSNIS PEQVYSLLLT NVIITGSTSL IEGMEQRIIK ELSIRFPQYK 420
421 LTTFANQVMM DRKIQGWLGA LTMANLPSWS LGKWYSKEDY ETLKRDRKQS QATNATN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP7 ARP9 HTL1 NFI1 NPL6 RSC1 RSC2 RSC3 RSC30 RSC4 RSC58 RSC6 RSC8 RSC9 RTT102 SFH1 SNF11 SNF12 SNF2 SNF5 SNF6 STH1 SWI1 SWI3 SWP82 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
ARP7 ARP9 RSC1 RSC2 RSC4 RSC6 RSC8 SAS5 SFH1 SNF11 SNF12 SNF2 SNF5 SNF6 STH1 SWI1 SWI3 TAF14 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP7 ARP9 ATG16 CSH1 GRS2 HTL1 MGR1 NFI1 NPL6 PCL9 RIM15 RSC1 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RSC9 RTT102 SFH1 STH1 URC2 YIL177C |
| View Details | Gavin AC, et al. (2006) |
|
ARP7 NPL6 RSC1 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RSC9 SFH1 STH1 |
| View Details | Ho Y, et al. (2002) |

|
ARP7 DIG1 DIG2 FET4 GFA1 GUS1 HAS1 HXT6 KSS1 MKT1 MSE1 NAP1 PHO84 PIM1 PMA1 PYC1 RGC1 RPA135 RPN10 SEN1 STE12 STE7 TEC1 UBI4 YDR239C YHR033W |
| View Details | Qiu et al. (2008) |

|
ARP7 ARP9 RSC1 RSC2 RSC4 RSC6 RSC8 RTT102 SFH1 SNF11 SNF12 SNF2 SNF5 SNF6 STH1 SWI1 SWI3 SWP82 TAF14 TFB5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | control 1 | McCusker D, et al (2007) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
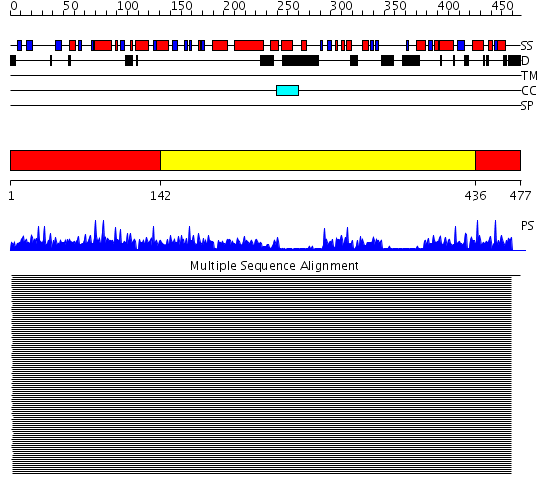
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] [436..477] |
1153.9794 | Actin | |
| 2 | View Details | [142..435] | 1153.9794 | Actin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)