













| General Information: |
|
| Name(s) found: |
DIS3 /
YOL021C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1001 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasmic exosome (RNase complex)
[IDA nuclear exosome (RNase complex) [IDA mitochondrion [IDA nuclear outer membrane [TAS |
| Biological Process: |
mRNA catabolic process
[IPI polyadenylation-dependent ncRNA catabolic process [IGI |
| Molecular Function: |
3'-5'-exoribonuclease activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVPAIAPRR KRLADGLSVT QKVFVRSRNG GATKIVREHY LRSDIPCLSR SCTKCPQIVV 60
61 PDAQNELPKF ILSDSPLELS APIGKHYVVL DTNVVLQAID LLENPNCFFD VIVPQIVLDE 120
121 VRNKSYPVYT RLRTLCRDSD DHKRFIVFHN EFSEHTFVER LPNETINDRN DRAIRKTCQW 180
181 YSEHLKPYDI NVVLVTNDRL NREAATKEVE SNIITKSLVQ YIELLPNADD IRDSIPQMDS 240
241 FDKDLERDTF SDFTFPEYYS TARVMGGLKN GVLYQGNIQI SEYNFLEGSV SLPRFSKPVL 300
301 IVGQKNLNRA FNGDQVIVEL LPQSEWKAPS SIVLDSEHFD VNDNPDIEAG DDDDNNESSS 360
361 NTTVISDKQR RLLAKDAMIA QRSKKIQPTA KVVYIQRRSW RQYVGQLAPS SVDPQSSSTQ 420
421 NVFVILMDKC LPKVRIRTRR AAELLDKRIV ISIDSWPTTH KYPLGHFVRD LGTIESAQAE 480
481 TEALLLEHDV EYRPFSKKVL ECLPAEGHDW KAPTKLDDPE AVSKDPLLTK RKDLRDKLIC 540
541 SIDPPGCVDI DDALHAKKLP NGNWEVGVHI ADVTHFVKPG TALDAEGAAR GTSVYLVDKR 600
601 IDMLPMLLGT DLCSLKPYVD RFAFSVIWEL DDSANIVNVN FMKSVIRSRE AFSYEQAQLR 660
661 IDDKTQNDEL TMGMRALLKL SVKLKQKRLE AGALNLASPE VKVHMDSETS DPNEVEIKKL 720
721 LATNSLVEEF MLLANISVAR KIYDAFPQTA MLRRHAAPPS TNFEILNEML NTRKNMSISL 780
781 ESSKALADSL DRCVDPEDPY FNTLVRIMST RCMMAAQYFY SGAYSYPDFR HYGLAVDIYT 840
841 HFTSPIRRYC DVVAHRQLAG AIGYEPLSLT HRDKNKMDMI CRNINRKHRN AQFAGRASIE 900
901 YYVGQVMRNN ESTETGYVIK VFNNGIVVLV PKFGVEGLIR LDNLTEDPNS AAFDEVEYKL 960
961 TFVPTNSDKP RDVYVFDKVE VQVRSVMDPI TSKRKAELLL K |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
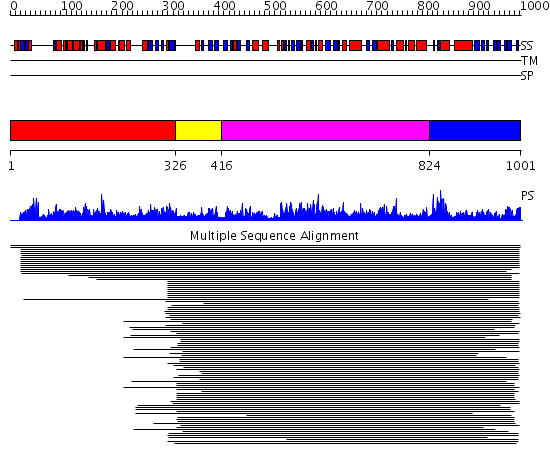
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..325] | 1.038904 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [326..415] | 1.081995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [416..823] | 168.39794 | RNB-like protein No confident structure predictions are available. | |
| 4 | View Details | [824..1001] | 14.39794 | C-terminal domain of RNA polymerase II subunit RBP4 (RpoE); N-terminal, heterodimerisation domain of RBP4 (RpoE) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)