













| General Information: |
|
| Name(s) found: |
ADE4 /
YMR300C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 510 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
purine nucleotide biosynthetic process
[TAS]
'de novo' IMP biosynthetic process [TAS] purine base metabolic process [IMP |
| Molecular Function: |
amidophosphoribosyltransferase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCGILGIVLA NQTTPVAPEL CDGCIFLQHR GQDAAGIATC GSRGRIYQCK GNGMARDVFT 60
61 QQRVSGLAGS MGIAHLRYPT AGSSANSEAQ PFYVNSPYGI NLAHNGNLVN TASLKRYMDE 120
121 DVHRHINTDS DSELLLNIFA AELEKHNKYR VNNEDVFHAL EGVYRLCRGG YACVGLLAGF 180
181 ALFGFRDPNG IRPLLFGERE NPDGTKDYML ASESVVFKAH NFTKYRDLKP GEAVIIPKNC 240
241 SKGEPEFKQV VPINSYRPDL FEYVYFARPD SVLDGISVYH TRLAMGSKLA ENILKQLKPE 300
301 DIDVVIPVPD TARTCALECA NVLGKPYREG FVKNRYVGRT FIMPNQRERV SSVRRKLNPM 360
361 ESEFKGKKVL IVDDSIVRGT TSKEIVNMAK ESGATKVYFA SAAPAIRYNH IYGIDLTDTK 420
421 NLIAYNRTDE EVAEVIGCER VIYQSLEDLI DCCKTDKITK FEDGVFTGNY VTGVEDGYIQ 480
481 ELEEKRESIA NNSSDMKAEV DIGLYNCADY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |
|
ADE4 ASC1 BFR2 BMS1 BRX1 BUD21 CBF5 CFT2 CKA1 CKA2 CKB1 CKB2 CMS1 CYS3 DBP8 DIA4 DIM1 DIP2 DIS3 ECM16 ECM29 EMG1 ENP1 ENP2 ERO1 ESF1 ESF2 FAP7 FIP1 GCN1 GLC7 HAS1 HCA4 HHF1, HHF2 HSL1 HTA2 IMP3 IMP4 KRE33 KRR1 LCP5 LTV1 MPP10 MRD1 MVD1 NAN1 NOB1 NOC2 NOC4 NOP1 NOP12 NOP14 NOP56 NOP58 NOP6 PAP1 PFS2 PNO1 PRE6 PRE9 PRP43 PTA1 PTI1 PWP2 RCL1 REF2 RML2 RNA1 RNA14 ROK1 RPP2B RRP12 RRP7 RRP9 RTG2 SAM1 SCL1 SEC21 SLX9 SOF1 SRO9 TIF1, TIF2 TSR1 UTP10 UTP11 UTP13 UTP15 UTP18 UTP20 UTP21 UTP22 UTP30 UTP4 UTP6 UTP7 UTP8 UTP9 YOR059C YSH1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
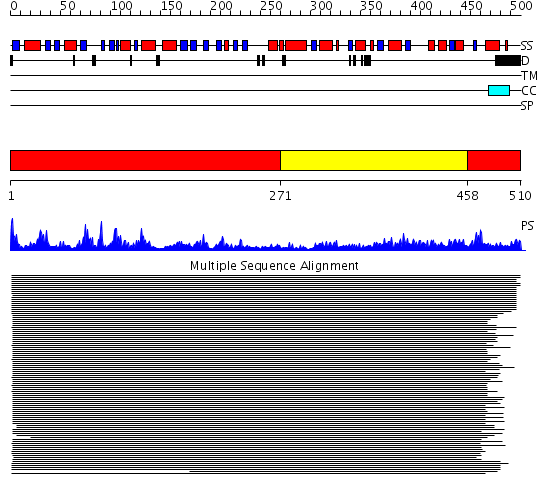
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..270] [458..510] |
1526.0 | Glutamine PRPP amidotransferase, C-terminal domain; Glutamine PRPP amidotransferase, N-terminal domain | |
| 2 | View Details | [271..457] | 1526.0 | Glutamine PRPP amidotransferase, C-terminal domain; Glutamine PRPP amidotransferase, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.61 |
Source: Reynolds et al. (2008)