













| General Information: |
|
| Name(s) found: |
RPL7B /
YPL198W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 244 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ribonucleoprotein complex
[IEA]
cytoplasm [IDA][IEA] cytosolic large ribosomal subunit [TAS] large ribosomal subunit [IEA] ribosome [IEA] nucleolus [IDA] intracellular [IEA] |
| Biological Process: |
translation
[TAS |
| Molecular Function: |
structural constituent of ribosome
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTEKILTPE SQLKKTKAQQ KTAEQIAAER AARKAANKEK RAIILERNAA YQKEYETAER 60
61 NIIQAKRDAK AAGSYYVEAQ HKLVFVVRIK GINKIPPKPR KVLQLLRLTR INSGTFVKVT 120
121 KATLELLKLI EPYVAYGYPS YSTIRQLVYK RGFGKINKQR VPLSDNAIIE ANLGKYGILS 180
181 IDDLIHEIIT VGPHFKQANN FLWPFKLSNP SGGWGVPRKF KHFIQGGSFG NREEFINKLV 240
241 KAMN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | control 1 | McCusker D, et al (2007) | |
| View Run | experimental2 - mih1-2 | McCusker D, et al (2007) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
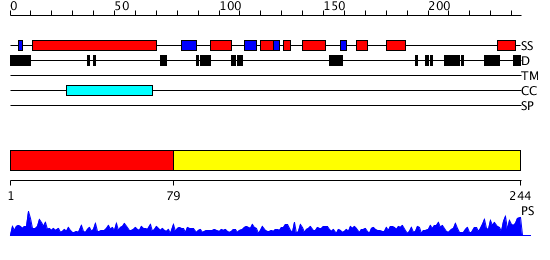
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | 26.677781 | No description for PF08079.3 was found. Confident ab initio structure predictions are available. | |
| 2 | View Details | [79..244] | 56.522879 | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. This file, 1S1I, Contains 60S subunit. The 40S Ribosomal Subunit Is In File 1S1H. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)