













| General Information: |
|
| Name(s) found: |
SWR1 /
YDR334W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1514 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Swr1 complex [IDA |
| Biological Process: |
chromatin remodeling
[IDA histone exchange [IMP |
| Molecular Function: |
structural molecule activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTSRKSHAK DKKAGGEQDL ADLKFRYDLL TNELFHLREF VSLVDYDPTH FNDSESFQKF 60
61 LRETHLSLEE RGEKFTDDVA KKGTNGDLTR RRRNLRTSTV VSSETTNEKK GDIELKLESI 120
121 APLVRNKCEE LKYKLSDHSN RKSIVPQKRP IQHLKKREAA KSLKFKSERK ENPLPLHEHI 180
181 AEERYDHIAK VEEPSEAFTI KCPSDDSSFE NTSEHYSDNF YFTTSSEEED IKKKRGRKKK 240
241 KPRIKLVVHP PKQTITNPLH VVKPGYESLH EYIASFKSLE DDLTLEEYNK YIDEQRRLLS 300
301 RLKKGIENGA LKYDKETDSL QPITSKEIKT IITYKPDPIS YFYKQQDLQI HTDHLINQGI 360
361 HMSKLFRSST KARIARAKKV SQMIEQHFKH VAGAEERKAK EEERHKKSLA RFAVQAVKKR 420
421 WNMAEKAYRI LRKDEEEQLK RIEGKQHLSK MLEKSTQLLE AQLNQVNDDG RSSTPSSDSN 480
481 DVLSESDDDM DDELSTSSDE DEEVDADVGL ENSPASTEAT PTDESLNLIQ LKEKYGHFNG 540
541 SSTVYDSRNK DEKFPTLDKH ESSSSESSVM TGEESSIYSS SENESQNEND RESDDKTPSV 600
601 GLSALFGKGE ESDGDLDLDD SEDFTVNSSS VEGEELEKDQ VDNSAATFER AGDFVHTQNE 660
661 NRDDIKDVEE DAETKVQEEQ LSVVDVPVPS LLRGNLRTYQ KQGLNWLASL YNNHTNGILA 720
721 DEMGLGKTIQ TISLLAYLAC EKENWGPHLI VVPTSVLLNW EMEFKRFAPG FKVLTYYGSP 780
781 QQRKEKRKGW NKPDAFHVCI VSYQLVVQDQ HSFKRKRWQY MVLDEAHNIK NFRSTRWQAL 840
841 LNFNTQRRLL LTGTPLQNNL AELWSLLYFL MPQTVIDGKK VSGFADLDAF QQWFGRPVDK 900
901 IIETGQNFGQ DKETKKTVAK LHQVLRPYLL RRLKADVEKQ MPAKYEHIVY CKLSKRQRFL 960
961 YDDFMSRAQT KATLASGNFM SIVNCLMQLR KVCNHPNLFE VRPILTSFVL EHCVASDYKD1020
1021 VERTLLKLFK KNNQVNRVDL DFLNLVFTLN DKDLTSYHAE EISKLTCVKN FVEEVNKLRE1080
1081 TNKQLQEEFG EASFLNFQDA NQYFKYSNKQ KLEGTVDMLN FLKMVNKLRC DRRPIFGKNL1140
1141 IDLLTKDRRV KYDKSSIIDN ELIKPLQTRV LDNRKIIDTF AVLTPSAVSL DMRKLALGLN1200
1201 DDSSVGENTR LKVMQNCFEV SNPLHQLQTK LTIAFPDKSL LQYDCGKLQK LAILLQQLKD1260
1261 NGHRALIFTQ MTKVLDVLEQ FLNYHGYLYM RLDGATKIED RQILTERFNT DSRITVFILS1320
1321 SRSGGLGINL TGADTVIFYD SDWNPAMDKQ CQDRCHRIGQ TRDVHIYRFV SEHTIESNIL1380
1381 KKANQKRQLD NVVIQEGDFT TDYFSKLSVR DLLGSELPEN ASGGDKPLIA DADVAAKDPR1440
1441 QLERLLAQAE DEDDVKAANL AMREVEIDND DFDESTEKKA ANEEEENHAE LDEYEGTAHV1500
1501 DEYMIRFIAN GYYY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
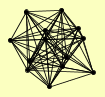
|
ARP4 ARP6 BDF1 RVB1 RVB2 SWC3 SWC4 SWC5 SWC7 SWR1 VPS71 VPS72 YAF9 |
| View Details | Hazbun TR, et al. (2003) |
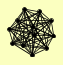
|
ARP4 EAF1 EAF5 EPL1 ESA1 RVB1 RVB2 SWC4 SWR1 TRA1 YAF9 YNG2 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP4 ARP5 ARP6 ARP8 CCA1 EAF1 EAF5 EAF6 EAF7 EPL1 ESA1 IES1 IES2 IES3 IES4 IES5 IES6 INO80 NHP10 NTO1 PIH1 RVB1 RVB2 SAS3 SWC3 SWC4 SWC5 SWC7 SWR1 TAH1 TRA1 VPS71 VPS72 YAF9 YNG2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
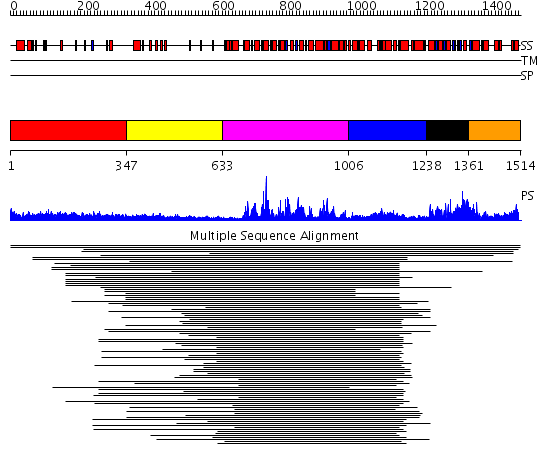
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..346] | 3.154902 | Heavy meromyosin subfragment | |
| 2 | View Details | [347..632] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [633..1005] | 114.004365 | SNF2 family N-terminal domain No confident structure predictions are available. | |
| 4 | View Details | [1006..1237] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1238..1360] | 22.031517 | Helicase conserved C-terminal domain No confident structure predictions are available. | |
| 6 | View Details | [1361..1514] | 4.202995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)