













| General Information: |
|
| Name(s) found: |
SIR3 /
YLR442C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 978 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear telomeric heterochromatin
[IDA mitochondrion [IDA nucleolus [IDA chromatin silencing complex [IDA nuclear telomere cap complex [IDA |
| Biological Process: |
loss of chromatin silencing involved in replicative cell aging
[IDA chromatin silencing [TAS double-strand break repair via nonhomologous end joining [IDA |
| Molecular Function: |
structural constituent of chromatin
[TAS histone binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKTLKDLDG WQVIITDDQG RVIDDNNRRR SRKRGGENVF LKRISDGLSF GKGESVIFND 60
61 NVTETYSVYL IHEIRLNTLN NVVEIWVFSY LRWFELKPKL YYEQFRPDLI KEDHPLEFYK 120
121 DKFFNEVNKS ELYLTAELSE IWLKDFIAVG QILPESQWND SSIDKIEDRD FLVRYACEPT 180
181 AEKFVPIDIF QIIRRVKEME PKQSDEYLKR VSVPVSGQKT NRQVMHKMGV ERSSKRLAKK 240
241 PSMKKIKIEP SADDDVNNGN IPSQRGTSTT HGSISPQEES VSPNISSASP SALTSPTDSS 300
301 KILQKRSISK ELIVSEEIPI NSSEQESDYE PNNETSVLSS KPGSKPEKTS TELVDGRENF 360
361 VYANNPEVSD DGGLEEETDE VSSESSDEAI IPVNKRRGAH GSELSSKIRK IHIQETQEFS 420
421 KNYTTETDNE MNGNGKPGIP RGNTKIHSMN ENPTPEKGNA KMIDFATLSK LKKKYQIILD 480
481 RFAPDNQVTD SSQLNKLTDE QSSLDVAGLE DKFRKACSSS GRETILSNFN ADINLEESIR 540
541 ESLQKRELLK SQVEDFTRIF LPIYDSLMSS QNKLFYITNA DDSTKFQLVN DVMDELITSS 600
601 ARKELPIFDY IHIDALELAG MDALYEKIWF AISKENLCGD ISLEALNFYI TNVPKAKKRK 660
661 TLILIQNPEN LLSEKILQYF EKWISSKNSK LSIICVGGHN VTIREQINIM PSLKAHFTEI 720
721 KLNKVDKNEL QQMIITRLKS LLKPFHVKVN DKKEMTIYNN IREGQNQKIP DNVIVINHKI 780
781 NNKITQLIAK NVANVSGSTE KAFKICEAAV EISKKDFVRK GGLQKGKLVV SQEMVPRYFS 840
841 EAINGFKDET ISKKIIGMSL LMRTFLYTLA QETEGTNRHT LALETVLIKM VKMLRDNPGY 900
901 KASKEIKKVI CGAWEPAITI EKLKQFSWIS VVNDLVGEKL VVVVLEEPSA SIMVELKLPL 960
961 EINYAFSMDE EFKNMDCI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
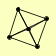
|
GPG1 SIR2 SIR3 SIR4 YLR278C |
| View Details | Ho Y, et al. (2002) |
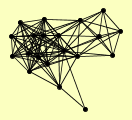
|
ADH6 BLM10 COR1 CYS4 GAS1 ILV5 RNR2 SAH1 SEC53 SES1 SIR1 SIR2 SIR3 SIR4 SRP1 TEF4 TFP1 TMA29 UBP8 YFL006W |
| View Details | Qiu et al. (2008) |
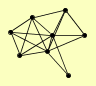
|
FPR4 NET1 PCC1 RIF1 SIR3 SIR4 SUB2 TBF1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
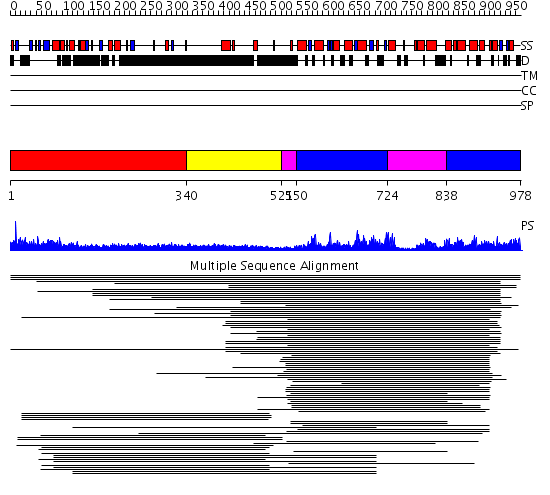
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..339] | 2.522879 | Heavy meromyosin subfragment | |
| 2 | View Details | [340..520] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [521..549] [724..837] |
57.154902 | CDC6, C-terminal domain; CDC6, N-domain | |
| 4 | View Details | [550..723] [838..978] |
57.154902 | CDC6, C-terminal domain; CDC6, N-domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)