













| General Information: |
|
| Name(s) found: |
TDH1 /
YJL052W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 332 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA fungal-type cell wall [IDA cytoplasm [IDA mitochondrion [IDA |
| Biological Process: |
gluconeogenesis
[IEP glycolysis [IEP |
| Molecular Function: |
glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIRIAINGFG RIGRLVLRLA LQRKDIEVVA VNDPFISNDY AAYMVKYDST HGRYKGTVSH 60
61 DDKHIIIDGV KIATYQERDP ANLPWGSLKI DVAVDSTGVF KELDTAQKHI DAGAKKVVIT 120
121 APSSSAPMFV VGVNHTKYTP DKKIVSNASC TTNCLAPLAK VINDAFGIEE GLMTTVHSMT 180
181 ATQKTVDGPS HKDWRGGRTA SGNIIPSSTG AAKAVGKVLP ELQGKLTGMA FRVPTVDVSV 240
241 VDLTVKLEKE ATYDQIKKAV KAAAEGPMKG VLGYTEDAVV SSDFLGDTHA SIFDASAGIQ 300
301 LSPKFVKLIS WYDNEYGYSA RVVDLIEYVA KA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ADH1 ADH2 ADH3 BMH2 BUD3 HSC82 HXK1 HYP2 RPL40B, RPL40A TDH1 TDH2 TRX2 |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Gavin AC, et al. (2006) |

|
ADE3 CSE1 DPS1 ILV5 IMD4 SAR1 TDH1 TOM1 YDR341C YNK1 |
| View Details | Qiu et al. (2008) |

|
TDH1 TDH2 TDH3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | control 1 | McCusker D, et al (2007) | |
| View Run | 3908: TAP-tagged, includes hir2(delta) in background | Green EM, et al (2005) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
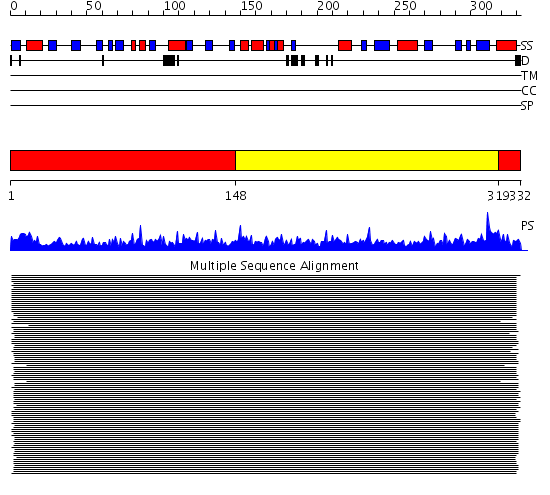
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] [319..332] |
1400.0 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | |
| 2 | View Details | [148..318] | 1400.0 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)