













| General Information: |
|
| Name(s) found: |
PSK1 /
YAL017W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1356 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
hexose metabolic process
[IGI protein amino acid phosphorylation [IMP |
| Molecular Function: |
protein serine/threonine kinase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPYIGASNLS EHSFVNLKEK HAITHKGTSS SVASLQTPPS PDQENHIDNE LENYDTSLSD 60
61 VSTPNKKEGD EFQQSLRDTF ASFRKTKPPP PLDFEQPRLP STASSSVDST VSSPLTDEDI 120
121 KELEFLPNES THSYSYNPLS PNSLAVRLRI LKRSLEIIIQ NPSMLLEPTP DDLPPLKEFA 180
181 GRRSSLPRTS ASANHLMNRN KSQIWNTTSA TLNAFVNNTS SSSAASSALS NKKPGTPVFP 240
241 NLDPTHSQTF HRANSLAYLP SILPEQDPLL KHNNSLFRGD YGNNISPERP SFRQPFKDQT 300
301 SNLRNSSLLN ERAYQEDETF LPHHGPSMDL LNEQRANLKS LLNLLNETLE KNTSERASDL 360
361 HMISLFNLNK LMLGDPKKNN SERDKRTEKL KKILLDSLAE PFFEHYNFIG DNPIADTDEL 420
421 KEEIDEFTGS GDTTAITDIR PQQDYGRILR TFTSTKNSAP QAIFTCSQED PWQFRAANDL 480
481 ACLVFGISQN AIRALTLMDL IHTDSRNFVL HKLLSTEGQE MVFTGEIIGI VQPETLSSSK 540
541 VVWASFWAKR KNGLLVCVFE KVPCDYVDVL LNLDDFGAEN IVDKCELLSD GPTLSSSSTL 600
601 SLPKMASSPT GSKLEYSLER KILEKSYTKP TSTENRNGDE NQLDGDSHSE PSLSSSPVRS 660
661 KKSVKFANDI KDVKSISQSL AKLMDDVRNG VVFDPDDDLL PMPIKVCNHI NETRYFTLNH 720
721 LSYNIPCAVS STVLEDELKL KIHSLPYQAG LFIVDSHTLD IVSSNKSILK NMFGYHFAEL 780
781 VGKSITEIIP SFPKFLQFIN DKYPALDITL HKNKGLVLTE HFFRKIQAEI MGDRKSFYTS 840
841 VGIDGLHRDG CEIKIDFQLR VMNSKVILLW VTHSRDVVFE EYNTNPSQLK MLKESELSLM 900
901 SSASSSASSS KKSSSRISTG TLKDMSNLST YEDLAHRTNK LKYEIGDDSR AHSQSTLSEQ 960
961 EQVPLENDKD SGEMMLADPE MKHKLELARI YSRDKSQFVK EGNFKVDENL IISKISLSPS1020
1021 TESLADSKSS GKGLSPLEEE KLIDENATEN GLAGSPKDED GIIMTNKRGN QPVSTFLRTP1080
1081 EKNIGAQKHV KKFSDFVSLQ KMGEGAYGKV NLCIHKKNRY IVVIKMIFKE RILVDTWVRD1140
1141 RKLGTIPSEI QIMATLNKKP HENILRLLDF FEDDDYYYIE TPVHGETGCI DLFDLIEFKT1200
1201 NMTEFEAKLI FKQVVAGIKH LHDQGIVHRD IKDENVIVDS KGFVKIIDFG SAAYVKSGPF1260
1261 DVFVGTIDYA APEVLGGNPY EGQPQDIWAI GILLYTVVFK ENPFYNIDEI LEGDLKFNNA1320
1321 EEVSEDCIEL IKSILNRCVP KRPTIDDINN DKWLVI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PSK1 PSK2 |
| View Details | Krogan NJ, et al. (2006) |
|
GET1 GET2 GET3 PSK1 TAT1 YHP1 |
| View Details | Gavin AC, et al. (2006) |
|
HEM13 PSK1 TAF7 VAS1 |
| View Details | Gavin AC, et al. (2002) |
|
ADA2 ADH1 BMH1 BMH2 CLU1 FUN19 GCN5 HEM13 HFI1 KAP114 KAP123 MED4 MES1 MNN4 MOT1 MYO1 MYO4 NGG1 NOP1 NTH1 NTH2 PFK2 PIK1 PSK1 PTC1 PUF6 RPB2 RPN9 RPO21 RPT5 RTG2 RVB2 SGF29 SGF73 SIN3 SIN4 SPT15 SPT20 SPT7 SPT8 TAF1 TAF10 TAF11 TAF12 TAF13 TAF14 TAF2 TAF3 TAF4 TAF5 TAF6 TAF7 TAF8 TAF9 TBF1 TEF4 UBP8 VAS1 VPS1 |
| View Details | Ho Y, et al. (2002) |

|
FUN30 GPH1 PSK1 PSK2 RVB1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
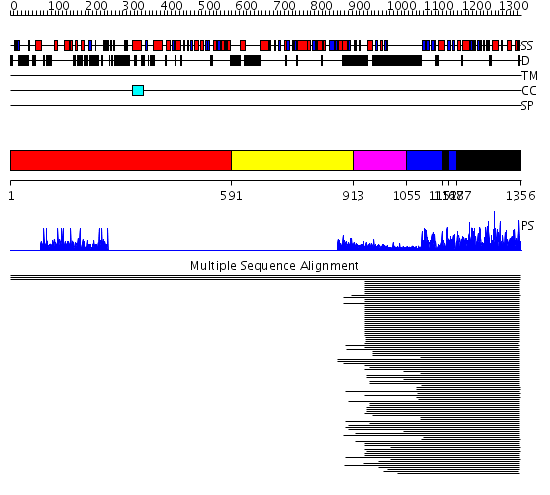
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..590] | 7.46 | Haloperoxidase (bromoperoxidase) | |
| 2 | View Details | [591..912] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [913..1054] | 12.045757 | C2 domain from protein kinase c (alpha) | |
| 4 | View Details | [1055..1151] [1167..1186] |
319.9897 | cAMP-dependent PK, catalytic subunit | |
| 5 | View Details | [1152..1166] [1187..1356] |
319.9897 | cAMP-dependent PK, catalytic subunit | |
| 6 | View Details | [906..1091] | 12.69897 | C2 domain from protein kinase c (alpha) | |
| 7 | View Details | [1092..1356] | 111.0 | No description for 2fh9A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)