













| General Information: |
|
| Name(s) found: |
SWI4 /
YER111C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1093 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
transcription
[IMP G1/S transition of mitotic cell cycle [IGI |
| Molecular Function: |
DNA binding
[IDA transcription factor activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPFDVLISNQ KDNTNHQNIT PISKSVLLAP HSNHPVIEIA TYSETDVYEC YIRGFETKIV 60
61 MRRTKDDWIN ITQVFKIAQF SKTKRTKILE KESNDMQHEK VQGGYGRFQG TWIPLDSAKF 120
121 LVNKYEIIDP VVNSILTFQF DPNNPPPKRS KNSILRKTSP GTKITSPSSY NKTPRKKNSS 180
181 SSTSATTTAA NKKGKKNASI NQPNPSPLQN LVFQTPQQFQ VNSSMNIMNN NDNHTTMNFN 240
241 NDTRHNLINN ISNNSNQSTI IQQQKSIHEN SFNNNYSATQ KPLQFFPIPT NLQNKNVALN 300
301 NPNNNDSNSY SHNIDNVINS SNNNNNGNNN NLIIVPDGPM QSQQQQQHHH EYLTNNFNHS 360
361 MMDSITNGNS KKRRKKLNQS NEQQFYNQQE KIQRHFKLMK QPLLWQSFQN PNDHHNEYCD 420
421 SNGSNNNNNT VASNGSSIEV FSSNENDNSM NMSSRSMTPF SAGNTSSQNK LENKMTDQEY 480
481 KQTILTILSS ERSSDVDQAL LATLYPAPKN FNINFEIDDQ GHTPLHWATA MANIPLIKML 540
541 ITLNANALQC NKLGFNCITK SIFYNNCYKE NAFDEIISIL KICLITPDVN GRLPFHYLIE 600
601 LSVNKSKNPM IIKSYMDSII LSLGQQDYNL LKICLNYQDN IGNTPLHLSA LNLNFEVYNR 660
661 LVYLGASTDI LNLDNESPAS IMNKFNTPAG GSNSRNNNTK ADRKLARNLP QKNYYQQQQQ 720
721 QQQPQNNVKI PKIIKTQHPD KEDSTADVNI AKTDSEVNES QYLHSNQPNS TNMNTIMEDL 780
781 SNINSFVTSS VIKDIKSTPS KILENSPILY RRRSQSISDE KEKAKDNENQ VEKKKDPLNS 840
841 VKTAMPSLES PSSLLPIQMS PLGKYSKPLS QQINKLNTKV SSLQRIMGEE IKNLDNEVVE 900
901 TESSISNNKK RLITIAHQIE DAFDSVSNKT PINSISDLQS RIKETSSKLN SEKQNFIQSL 960
961 EKSQALKLAT IVQDEESKVD MNTNSSSHPE KQEDEEPIPK STSETSSPKN TKADAKFSNT1020
1021 VQESYDVNET LRLATELTIL QFKRRMTTLK ISEAKSKINS SVKLDKYRNL IGITIENIDS1080
1081 KLDDIEKDLR ANA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
SWI4 SWI6 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
SWI4 SWI6 |
| View Details | Krogan NJ, et al. (2006) |
|
BUD2 MBP1 MSA2 SWI4 SWI6 WHI5 |
| View Details | Gavin AC, et al. (2006) |

|
SWI4 SWI6 |
| View Details | Ho Y, et al. (2002) |
|
ASF1 CDC13 EDE1 KAP95 PTC2 RAD53 SRP1 SWI4 TBF1 UTP22 YTA7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
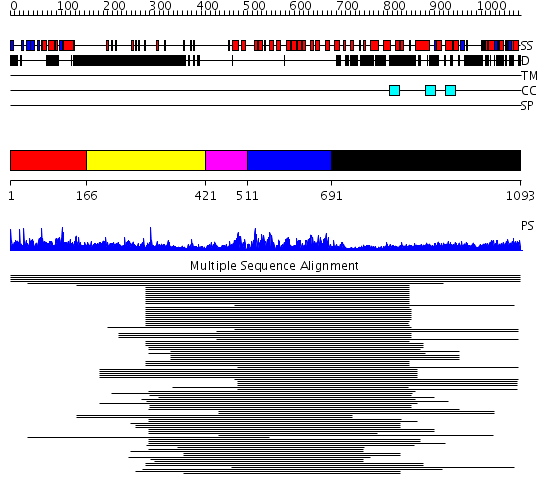
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 259.38764 | DNA-binding domain of Mlu1-box binding protein MBP1 | |
| 2 | View Details | [166..420] | 3.381996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [421..510] | 26.154902 | GA bindinig protein (GABP) beta 1 | |
| 4 | View Details | [511..690] | 127.457575 | Swi6 ankyrin-repeat fragment | |
| 5 | View Details | [691..1093] | 30.801997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)