













| General Information: |
|
| Name(s) found: |
SLA2 /
YNL243W
[SGD]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 968 amino acids |
Gene Ontology: |
|
| Cellular Component: |
incipient cellular bud site
[IDA actin cortical patch [IDA |
| Biological Process: |
bipolar cellular bud site selection
[IMP cellular cell wall organization [IMP telomere maintenance [IMP actin filament organization [IMP endocytosis [IMP exocytosis [IMP |
| Molecular Function: |
protein binding, bridging
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRIDSDLQK ALKKACSVEE TAPKRKHVRA CIVYTWDHQS SKAVFTTLKT LPLANDEVQL 60
61 FKMLIVLHKI IQEGHPSALA EAIRDRDWIR SLGRVHSGGS SYSKLIREYV RYLVLKLDFH 120
121 AHHRGFNNGT FEYEEYVSLV SVSDPDEGYE TILDLMSLQD SLDEFSQIIF ASIQSERRNT 180
181 ECKISALIPL IAESYGIYKF ITSMLRAMHR QLNDAEGDAA LQPLKERYEL QHARLFEFYA 240
241 DCSSVKYLTT LVTIPKLPVD APDVFLINDV DESKEIKFKK REPSVTPART PARTPTPTPP 300
301 VVAEPAISPR PVSQRTTSTP TGYLQTMPTG ATTGMMIPTA TGARNAIFPQ ATAQMQPDFW 360
361 ANQQAQFANE QNRLEQERVQ QLQQQQAQQE LFQQQLQKAQ QDMMNMQLQQ QNQHQNDLIA 420
421 LTNQYEKDQA LLQQYDQRVQ QLESEITTMD STASKQLANK DEQLTALQDQ LDVWERKYES 480
481 LAKLYSQLRQ EHLNLLPRFK KLQLKVNSAQ ESIQKKEQLE HKLKQKDLQM AELVKDRDRA 540
541 RLELERSINN AEADSAAATA AAETMTQDKM NPILDAILES GINTIQESVY NLDSPLSWSG 600
601 PLTPPTFLLS LLESTSENAT EFATSFNNLI VDGLAHGDQT EVIHCVSDFS TSMATLVTNS 660
661 KAYAVTTLPQ EQSDQILTLV KRCAREAQYF FEDLMSENLN QVGDEEKTDI VINANVDMQE 720
721 KLQELSLAIE PLLNIQSVKS NKETNPHSEL VATADKIVKS SEHLRVDVPK PLLSLALMII 780
781 DAVVALVKAA IQCQNEIATT TSIPLNQFYL KNSRWTEGLI SAAKAVAGAT NVLITTASKL 840
841 ITSEDNENTS PEQFIVASKE VAASTIQLVA ASRVKTSIHS KAQDKLEHCS KDVTDACRSL 900
901 GNHVMGMIED DHSTSQQQQP LDFTSEHTLK TAEMEQQVEI LKLEQSLSNA RKRLGEIRRH 960
961 AYYNQDDD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
LSB3 SLA2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
ABP1 AIP1 ARC40 ARP2 BEM1 BUD6 CAP1 CAP2 COF1 CRN1 DLD2 GLK1 LAS17 OYE2 PAN1 PFY1 RVS167 SAC6 SLA1 SLA2 SRV2 TPM1 TPM2 TWF1 VRP1 |
| View Details | Gavin AC, et al. (2006) |

|
END3 LSB3 SLA2 |
| View Details | Gavin AC, et al. (2002) |
|
BSP1 BZZ1 CHC1 ECM25 EDE1 END3 INP52 LAS17 LSB3 RAD51 SLA1 SLA2 STM1 SYP1 TFP1 VRP1 YDR348C YPT7 YSC84 |
| View Details | Qiu et al. (2008) |

|
ABP1 ABP140 ACT1 AIP1 ARP2 BEM1 BNI1 BNI4 BSP1 BUD6 CAP1 CAP2 COF1 CRN1 EDE1 END3 ENT1 ENT2 ENT3 GLK1 GYL1 INP53 IQG1 LAS17 MYO5 PAN1 PFY1 PXL1 RVS161 RVS167 SAC6 SCD5 SCP1 SLA1 SLA2 SPA2 SRV2 TPM1 TPM2 TWF1 VRP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Drees BL, et al. (2001) | ||
| View Screen | 2 | Drees BL, et al. (2001) | ||
| View Screen | 2 | Drees BL, et al. (2001) | ||
| View Screen | 2 | Drees BL, et al. (2001) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
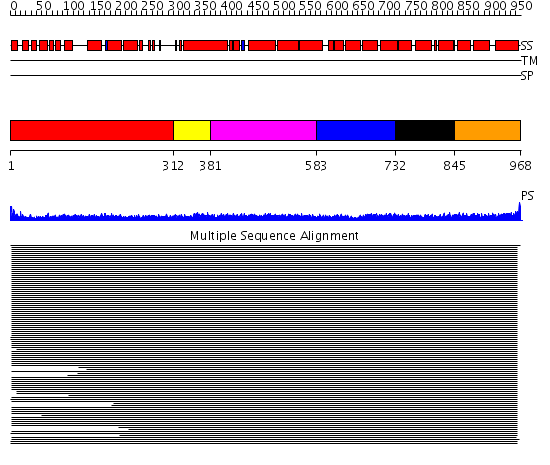
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..311] | 16.30103 | Heavy meromyosin subfragment | |
| 2 | View Details | [312..380] | 8.69897 | Tropomyosin | |
| 3 | View Details | [381..582] | 8.69897 | Tropomyosin | |
| 4 | View Details | [583..731] | 11.77 | alpha-actinin | |
| 5 | View Details | [732..844] | 11.77 | alpha-actinin | |
| 6 | View Details | [845..968] | 11.77 | alpha-actinin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)