













| General Information: |
|
| Name(s) found: |
OYE2 /
YHR179W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 400 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA mitochondrion [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
NADPH dehydrogenase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPFVKDFKPQ ALGDTNLFKP IKIGNNELLH RAVIPPLTRM RAQHPGNIPN RDWAVEYYAQ 60
61 RAQRPGTLII TEGTFPSPQS GGYDNAPGIW SEEQIKEWTK IFKAIHENKS FAWVQLWVLG 120
121 WAAFPDTLAR DGLRYDSASD NVYMNAEQEE KAKKANNPQH SITKDEIKQY VKEYVQAAKN 180
181 SIAAGADGVE IHSANGYLLN QFLDPHSNNR TDEYGGSIEN RARFTLEVVD AVVDAIGPEK 240
241 VGLRLSPYGV FNSMSGGAET GIVAQYAYVL GELERRAKAG KRLAFVHLVE PRVTNPFLTE 300
301 GEGEYNGGSN KFAYSIWKGP IIRAGNFALH PEVVREEVKD PRTLIGYGRF FISNPDLVDR 360
361 LEKGLPLNKY DRDTFYKMSA EGYIDYPTYE EALKLGWDKN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
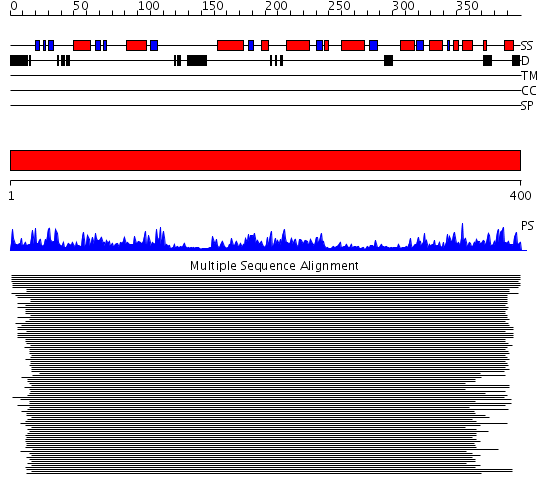
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..400] | 10123.0 | Old yellow enzyme (OYE) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)