













| General Information: |
|
| Name(s) found: |
RPN1 /
YHR027C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 993 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA proteasome regulatory particle, base subcomplex [IPI cytoplasm [IDA endoplasmic reticulum [IDA |
| Biological Process: |
ubiquitin-dependent protein catabolic process
[TAS |
| Molecular Function: |
endopeptidase activity
[TAS protein binding, bridging [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDESDKKQQ TIDEQSQISP EKQTPNKKDK KKEEEEQLSE EDAKLKTDLE LLVERLKEDD 60
61 SSLYEASLNA LKESIKNSTS SMTAVPKPLK FLRPTYPDLC SIYDKWTDPN LKSSLADVLS 120
121 ILAMTYSENG KHDSLRYRLL SDVSDFEGWG HEYIRHLALE IGEVYNDQVE KDAEDETSSD 180
181 GSKSDGSAAT SGFEFSKEDT LRLCLDIVPY FLKHNGEEDA VDLLLEIESI DKLPQFVDEN 240
241 TFQRVCQYMV ACVPLLPPPE DVAFLKTAYS IYLSQNELTD AIALAVRLGE EDMIRSVFDA 300
301 TSDPVMHKQL AYILAAQKTS FEYEGVQDII GNGKLSEHFL YLAKELNLTG PKVPEDIYKS 360
361 HLDNSKSVFS SAGLDSAQQN LASSFVNGFL NLGYCNDKLI VDNDNWVYKT KGDGMTSAVA 420
421 SIGSIYQWNL DGLQQLDKYL YVDEPEVKAG ALLGIGISAS GVHDGEVEPA LLLLQDYVTN 480
481 PDTKISSAAI LGLGIAFAGS KNDEVLGLLL PIAASTDLPI ETAAMASLAL AHVFVGTCNG 540
541 DITTSIMDNF LERTAIELKT DWVRFLALAL GILYMGQGEQ VDDVLETISA IEHPMTSAIE 600
601 VLVGSCAYTG TGDVLLIQDL LHRLTPKNVK GEEDADEEET AEGQTNSISD FLGEQVNEPT 660
661 KNEEAEIEVD EMEVDAEGEE VEVKAEITEK KNGESLEGEE IKSEEKKGKS SDKDATTDGK 720
721 NDDEEEEKEA GIVDELAYAV LGIALIALGE DIGKEMSLRH FGHLMHYGNE HIRRMVPLAM 780
781 GIVSVSDPQM KVFDTLTRFS HDADLEVSMN SIFAMGLCGA GTNNARLAQL LRQLASYYSR 840
841 EQDALFITRL AQGLLHLGKG TMTMDVFNDA HVLNKVTLAS ILTTAVGLVS PSFMLKHHQL 900
901 FYMLNAGIRP KFILALNDEG EPIKVNVRVG QAVETVGQAG RPKKITGWIT QSTPVLLNHG 960
961 ERAELETDEY ISYTSHIEGV VILKKNPDYR EEE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
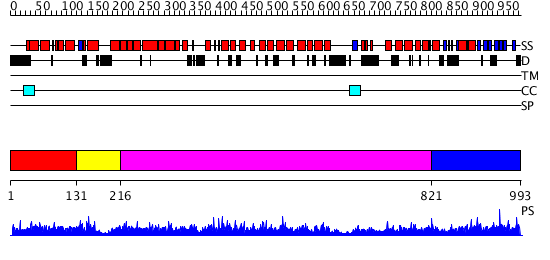
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..130] | 2.067963 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [131..215] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [216..820] | 2.08 | No description for 2ie3A was found. | |
| 4 | View Details | [821..993] | 3.08298 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)