













| General Information: |
|
| Name(s) found: |
MET18 /
YIL128W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1032 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleoplasm
[IMP |
| Biological Process: |
telomere maintenance
[IMP nucleotide-excision repair [IMP transcription from RNA polymerase II promoter [IGI methionine metabolic process [IMP |
| Molecular Function: |
RNA polymerase II transcription factor activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTPDELNSAV VTFMANLNID DSKANETAST VTDSIVHRSI KLLEVVVALK DYFLSENEVE 60
61 RKKALTCLTT ILAKTPKDHL SKNECSVIFQ FYQSKLDDQA LAKEVLEGFA ALAPMKYVSI 120
121 NEIAQLLRLL LDNYQQGQHL ASTRLWPFKI LRKIFDRFFV NGSSTEQVKR INDLFIETFL 180
181 HVANGEKDPR NLLLSFALNK SITSSLQNVE NFKEDLFDVL FCYFPITFKP PKHDPYKISN 240
241 QDLKTALRSA ITATPLFAED AYSNLLDKLT ASSPVVKNDT LLTLLECVRK FGGSSILENW 300
301 TLLWNALKFE IMQNSEGNEN TLLNPYNKDQ QSDDVGQYTN YDACLKIINL MALQLYNFDK 360
361 VSFEKFFTHV LDELKPNFKY EKDLKQTCQI LSAIGSGNVE IFNKVISSTF PLFLINTSEV 420
421 AKLKLLIMNF SFFVDSYIDL FGRTSKESLG TPVPNNKMAE YKDEIIMILS MALTRSSKAE 480
481 VTIRTLSVIQ FTKMIKMKGF LTPEEVSLII QYFTEEILTD NNKNIYYACL EGLKTISEIY 540
541 EDLVFEISLK KLLDLLPDCF EEKIRVNDEE NIHIETILKI ILDFTTSRHI LVKESITFLA 600
601 TKLNRVAKIS KSREYCFLLI STIYSLFNNN NQNENVLNEE DALALKNAIE PKLFEIITQE 660
661 SAIVSDNYNL TLLSNVLFFT NLKIPQAAHQ EELDRYNELF ISEGKIRILD TPNVLAISYA 720
721 KILSALNKNC QFPQKFTVLF GTVQLLKKHA PRMTETEKLG YLELLLVLSN KFVSEKDVIG 780
781 LFDWKDLSVI NLEVMVWLTK GLIMQNSLES SEIAKKFIDL LSNEEIGSLV SKLFEVFVMD 840
841 ISSLKKFKGI SWNNNVKILY KQKFFGDIFQ TLVSNYKNTV DMTIKCNYLT ALSLVLKHTP 900
901 SQSVGPFIND LFPLLLQALD MPDPEVRVSA LETLKDTTDK HHTLITEHVS TIVPLLLSLS 960
961 LPHKYNSVSV RLIALQLLEM ITTVVPLNYC LSYQDDVLSA LIPVLSDKKR IIRKQCVDTR1020
1021 QVYYELGQIP FE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CIA1 MET18 RAD3 |
| View Details | Krogan NJ, et al. (2006) |
|
CIA1 MET18 YHR122W |
| View Details | Gavin AC, et al. (2006) |
|
MET18 RAD3 |
| View Details | Gavin AC, et al. (2002) |
|
ASC1 CLU1 ECM29 GCN1 GCN20 MET18 RAD3 RPN10 RPT3 RPT6 TFP1 TUB3 YDJ1 |
| View Details | Ho Y, et al. (2002) |
|
AAC1 AAC3 ACC1 ACH1 ACO1 ADH2 ADR1 ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 ATG12 ATP3 BCY1 BNI1 CCL1 CIA1 CRM1 DED1 DOG1 DOP1 DUR1,2 ECM29 FAA4 FET3 GCD1 GCD11 GCD2 GCD6 GCD7 GCN3 GFA1 GLT1 GND1 GUS1 GYP5 HOR2 HXT6 IDH1 IDH2 ILV5 IMH1 ISA2 KAP122 KGD1 KIN28 LOS1 LSC1 LYS12 MCM4 MCM7 MDH1 MDH2 MDY2 MET10 MET18 NGL2 NMD5 PFK1 PHO84 PRB1 PRE10 PRO3 PYC1 PYC2 RAD3 RET1 RHR2 RPA135 RPA190 RPA49 RPB5 RPC19 RPC25 RPC34 RPC40 RPC82 RPN1 RPN3 RPN8 RPO26 RPO31 RPT3 RPT4 RVS167 SCW4 SEC27 SPC24 SPC25 SRY1 SUI3 TBS1 TFB3 TFP1 THI22 TRP3 TSA2 UBP9 URA7 VAC8 VMA8 YBR184W YER138C YFL042C YHR112C YJR029W YLR243W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
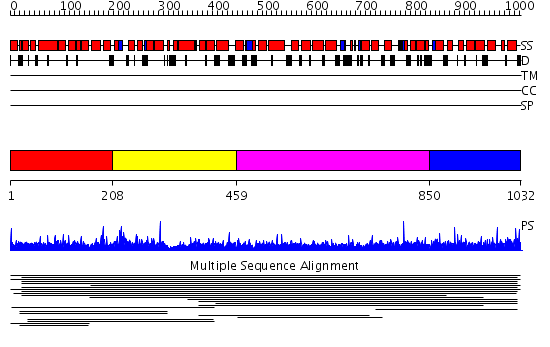
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..207] | 2.010957 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [208..458] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [459..849] | 1.013918 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [850..1032] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)