













| General Information: |
|
| Name(s) found: |
YDR333C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 723 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSRALRRLQ DDNALLESLL SNSNANKMTS GKSTAGNIQK RENIFSMMNN VRDSDNSTDE 60
61 GQMSEQDEEA AAAGERDTQS NGQPKRITLA SKSSRRKKNK KAKRKQKNHT AEAAKDKGSD 120
121 DDDDDEEFDK IIQQFKKTDI LKYGKTKNDD TNEEGFFTAS EPEEASSQPW KSFLSLESDP 180
181 GFTKFPISCL RHSCKFFQND FKKLDPHTEF KLLFDDISPE SLEDIDSMTS TPVSPQQLKQ 240
241 IQRLKRLIRN WGGKDHRLAP NGPGMHPQHL KFTKIRDDWI PTQRGELSMK LLSSDDLLDW 300
301 QLWERPLDWK DVIQNDVSQW QKFISFYKFE PLNSDLSKKS MMDFYLSVIV HPDHEALINL 360
361 ISSKFPYHVP GLLQVALIFI RQGDRSNTNG LLQRALFVFD RALKANIIFD SLNCQLPYIY 420
421 FFNRQFYLAI FRYIQSLAQR GVIGTASEWT KVLWSLSPLE DPLGCRYFLD HYFLLNNDYQ 480
481 YIIELSNSPL MNCYKQWNTL GFSLAVVLSF LRINEMSSAR NALLKAFKHH PLQLSELFKE 540
541 KLLGDHALTK DLSIDGHSAE NLELKAYMAR FPLLWNRNEE VTFLHDEMSS ILQDYHRGNV 600
601 TIDSNDGQDH NNINNLQSPF FIAGIPINLL RFAILSEESS VMAAIPSFIW SDNEVYEFDV 660
661 LPPMPTSKES IEVVENIKTF INEKDLAVLQ AERMQDEDLL NQIRQISLQQ YIHENEESNE 720
721 NEG |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
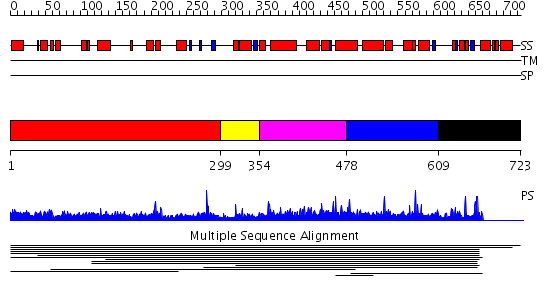
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..298] | 1.008954 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [299..353] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [354..477] | 9.73 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 4 | View Details | [478..608] | 9.73 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 5 | View Details | [609..723] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)