













| General Information: |
|
| Name(s) found: |
TAX4 /
YJL083W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 604 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
cellular cell wall organization
[IGI inositol lipid-mediated signaling [IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHFPKKKHSG NLSVVELPKE ALQDSLTAAQ ITFKRYAHPN GNAGSAERPR HLKVESAPVV 60
61 KSEPSLPRMR QPEPRSINHQ YSRETLPGHS EAFSVPTTPL QTIHYDVRNK ASNSPSSIAA 120
121 AETAAYLAHT NSFSNRSSGV GSRDPVMDTE TKPPRAPSAL KNELQLNRMR IPPPSYDNNV 180
181 RSRSISPQVS YSTSLSSSCS ISSDGEETSY REKSTDEAFP PEPSMSSYSL ASKASAKASL 240
241 TDPSQRQQES DYTAMNKLNG GNIIYKGTLP DLIPRSQRKT SKPRFKHRLL RSPEQQQENL 300
301 SRVYSDQTQN GRAIINTQQN VKLKTTMRRG KYAITDNDET FPYDRKSVSS DSDTDEDSNV 360
361 MEIKDKKKKS RRSKIKKGLK TTAAVVGSST SVLPFPHHHH HHHQLHNPNS HHLHTHHHTS 420
421 SHKFNEDKPW KSHRDLGFIT EQERKRYESM WVSNRYSYLR LLPWWPSLAN EDDESHLQPL 480
481 NLPQDGLMLN LVVKDIWYRS NLPRDLLVQI YNMVDTRKDG TLDRKSFIVG MWLVDQCLYG 540
541 RKLTNELDQR VWNSVDGYVL GTINVKPATS DHYHNANNPL DKPSKLSVRQ ELKNIKRDLR 600
601 NVRI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
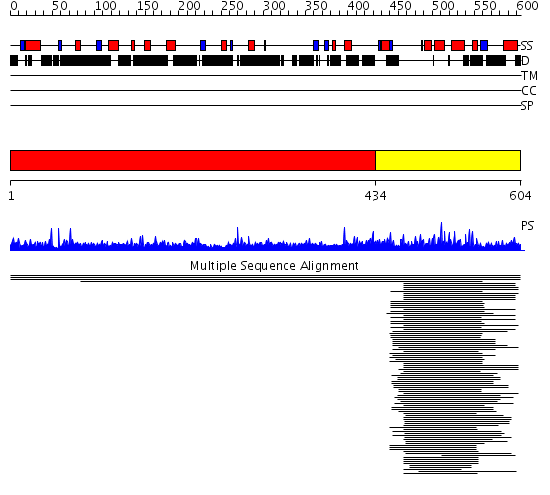
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..433] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [434..604] | 56.274545 | Eps15 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)