













| General Information: |
|
| Name(s) found: |
ROM1 /
YGR070W
[SGD]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1155 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[TAS |
| Biological Process: |
small GTPase mediated signal transduction
[IPI cellular cell wall organization [IPI actin filament organization [TAS budding cell bud growth [IPI |
| Molecular Function: |
Rho guanyl-nucleotide exchange factor activity
[IMP signal transducer activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSNELDLRN KYFYEIFGKK RKSDTSTPTQ LFSGSKVQTN INEISITNDE DEDSTEDENK 60
61 ASLKDYTLGH DTGARYRIAP DCSSHQLKAS PVLHISTNLN SSPQSFTGDQ ISPTNKKISI 120
121 NDSTRQDKGN SCTTTSSPSQ KRSNVLLPHV RKHSSPSLLS FSKNSGSHMG DPNQLSTPPT 180
181 PKSAGHTMEL HSSFNGKHSS SSTSSLFALE SLKTQNRRSS NSSNHSSQYR RHTNQHQRHH 240
241 SRSKSSPVSL TEISMIKGTP LVYPALLSLI AIKFKQTIKL STHKKMGLLY RDSFTGKQAI 300
301 DTLCLIIGSL DRNLGMLIGK SLEAQKLFHD VLYDHGVRDS VLEIYELSSE SIFMAHQSQS 360
361 STSIANTFSS SSSSVNSLRT KTEIYGVFVP LTHCYSSTCS LEKLCYSISC PNRLQQQANL 420
421 HLKLGGGLKR NISLALDKED DERISWTNSV PKSVWESLSK QQIKRQEAIY ELFTTEKKFV 480
481 KSLEIIRDTF MKKLLETNII PSDVRINFVK HVFAHINEIY SVNREFLKAL AQRQSLSPIC 540
541 PGIADIFLQY LPFFDPFLSY IASRPYAKYL IETQRSVNPN FARFDDEVSN SSLRHGIDSF 600
601 LSQGVSRPGR YSLLVREIIH FSDPVTDKDD LQMLMKVQDL LKDLMKRIDR ASGAAQDRYD 660
661 VKVLKQKILF KNEYVNLGLN NEKRKIKHEG LLSRKDVNKT DASFSGDIQF YLLDNMLLFL 720
721 KSKAVNKWHQ HTVFQRPIPL PLLFICPAED MPPIKRYVTE NPNCSAGVLL PQYQTSNPKN 780
781 AIVFAYYGTK QQYQVTLYAP QPAGLQTLIE KVKQEQKRLL DETKHITFKQ MVGQFFHSYI 840
841 NTNRVNDVLI CHAGKILLVA TNMGLFVLNY ATSINQKPVH LLHKISISQI SVLEEYKVMI 900
901 LLIDKKLYGC PLDVIDDAEN ADFLFRKNSK VLFKYVAMFK DGFCNGKRII MIAHHFLHAA 960
961 QLLIVNPLIF DFNSGNFKKN LKAGLVDFSV DSEPLSFSFL ENKICIGCKK NIKILNVPEV1020
1021 CDKNGFKMRE LLNLHDNKVL ANMYKETFKV VSMFPIKNST FACFPELCFF LNKQGKREET1080
1081 KGCFHWEGEP EQFACSYPYI VAINSNFIEI RHIENGELVR CVLGNKIRML KSYAKKILYC1140
1141 YEDPQGFEII ELLNF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
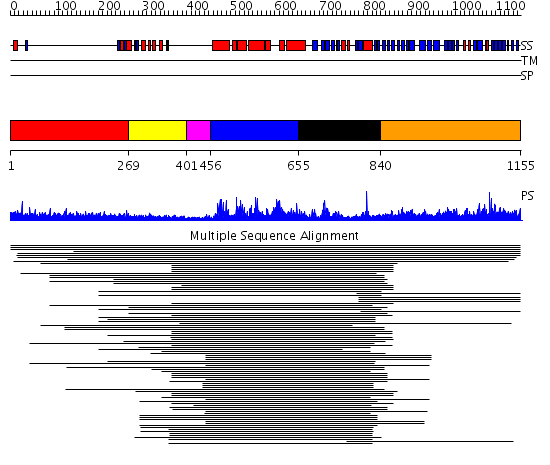
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..268] | 12.1 | RBP1 | |
| 2 | View Details | [269..400] | 16.721246 | Domain found in Dishevelled, Egl-10, and Pleckstrin No confident structure predictions are available. | |
| 3 | View Details | [401..455] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [456..654] | 160.927757 | GEF of intersectin | |
| 5 | View Details | [655..839] | 160.927757 | GEF of intersectin | |
| 6 | View Details | [840..1155] | 89.60206 | CNH domain No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)