













| General Information: |
|
| Name(s) found: |
MAK5 /
YBR142W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 773 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
ribosome biogenesis
[RCA rRNA processing [TAS ribosomal large subunit assembly [TAS |
| Molecular Function: |
ATP-dependent RNA helicase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKKRAPQKG KTVTKPQEII VDESKLNWKP VDIPDTLDDF GGFYGLEEID GVDVKVVDGK 60
61 VTFVTKKDSK VLKDSNKEKV GDDQESVENE SGSDSESELL EFKNLDDIKE GELSAASYSS 120
121 SDEDEQGNIE SSKLTDPSED VDEDVDEDVL KENVFNKDIN IDDISPVNLP EWTNLAPLSM 180
181 TILQSLQNLN FLRPTEIQKK SIPVIMQGVD VMGKASTGSG KTLAYGIPIV EKLISNFSQK 240
241 NKKPISLIFT PTRELAHQVT DHLKKICEPV LAKSQYSILS LTGGLSIQKQ QRLLKYDNSG 300
301 QIVIATPGRF LELLEKDNTL IKRFSKVNTL ILDEADRLLQ DGHFDEFEKI IKHLLVERRK 360
361 NRENSEGSSK IWQTLIFSAT FSIDLFDKLS SSRQVKDRRF KNNEDELNAV IQHLMSKIHF 420
421 NSKPVIIDTN PESKVSSQIK ESLIECPPLE RDLYCYYFLT MFPGTTLIFC NAIDSVKKLT 480
481 VYLNNLGIPA FQIHSSMTQK NRLKSLERFK QQSAKQKTIN HSNPDSVQLS TVLIASDVAA 540
541 RGLDIPGVQH VIHYHLPRST DIYIHRSGRT ARAGSEGVSA MICSPQESMG PLRKLRKTLA 600
601 TKNSVSTDLN SRSTNRKPIK WQNTVPLLPI ETDILSQLRE RSRLAGELAD HEIASNSLRK 660
661 DDNWLKKAAD ELGIDVDSDE DDISKSNSDT FLLKNKNKKM QKTINKDKVK AMRATLNELL 720
721 SVPIRKDRRQ KYLTGGLVNL ADNLVKKRGH NSIIGHEKTN ALETLKKKKK RNN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
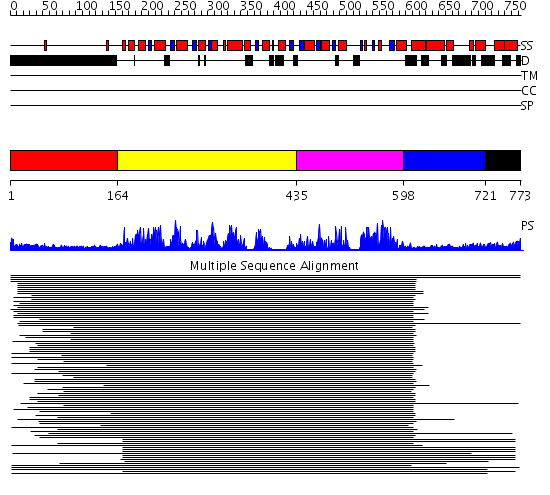
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..163] | 4.338997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [164..434] | 461.228787 | Putative DEAD box RNA helicase | |
| 3 | View Details | [435..597] | 461.228787 | Putative DEAD box RNA helicase | |
| 4 | View Details | [598..720] | 9.0 | Nucleotide excision repair enzyme UvrB | |
| 5 | View Details | [721..773] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)