













| General Information: |
|
| Name(s) found: |
NOP12 /
YOL041C
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 459 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
rRNA metabolic process
[IMP |
| Molecular Function: |
RNA binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSAIDNLFG NIDEKKIESS VDKLFSSSCG PINKLEVKSK TRTVLPDSKK RERAAEADQE 60
61 EKEASKPDVS DEQTEEVALP KVKKAKKSKR NDEDEDLEAR YYAKLLNEEA EAEDDKPTVT 120
121 KTDETSVPLT SAAKKVDFKE DELEKAERTV FIGNILSTVI TSKKVYKEFK KLFGTNPIAE 180
181 TEESGNEKEE ESSKKSDNNE FAIESIRFRS ISFDEALPRK VAFVQQKFHK SRDTINAYIV 240
241 YKNKSAVRKI CSNLNAVVFQ DHHLRVDSVA HPAPHDKKRS IFVGNLDFEE IEESLWKHFE 300
301 PCGDIEYVRI IRDSKTNMGK GFAYVQFKDL QSVNKALLLN EKPMKSQKQE DENTKKPTKK 360
361 ARKLRVSRCK NMKKGTTIGT GLDRNGLTDS QRTRAGRAKK ILGKADRATL GQEITIEGLR 420
421 AKKGEGSTHL KKKKQRSATG RVTKRSIAFK KAQAEKSKK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | #35 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
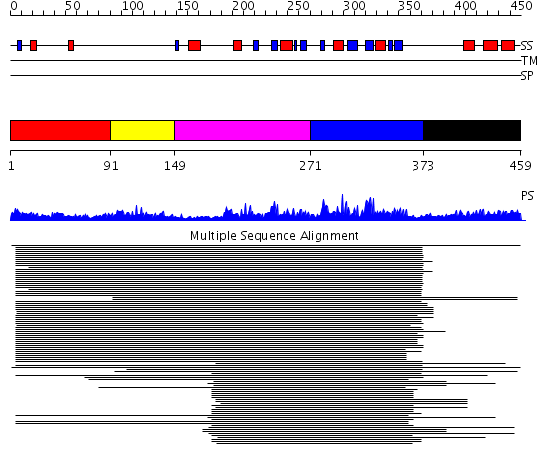
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 7.69897 | Nucleolin | |
| 2 | View Details | [91..148] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [149..270] | 109.519517 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 4 | View Details | [271..372] | 109.519517 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 5 | View Details | [373..459] | 1.362997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)