













| General Information: |
|
| Name(s) found: |
SKP1 /
YDR328C
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 194 amino acids |
Gene Ontology: |
|
| Cellular Component: |
RAVE complex
[IPI nucleus [IDA cytoplasm [IDA CBF3 complex [IDA SCF ubiquitin ligase complex [IDA kinetochore [TAS |
| Biological Process: |
G2/M transition of mitotic cell cycle
[IMP protein complex assembly [IDA regulation of exit from mitosis [IMP G1/S transition of mitotic cell cycle [IMP cytokinesis [IMP SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [IDA regulation of protein complex assembly [IPI protein neddylation [IGI kinetochore assembly [IDA vacuolar acidification [IGI protein ubiquitination during ubiquitin-dependent protein catabolic process [IDA |
| Molecular Function: |
DNA replication origin binding
[IPI ubiquitin-protein ligase activity [IDA protein binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVTSNVVLVS GEGERFTVDK KIAERSLLLK NYLNDMHDSN LQNNSDSESD SDSETNHKSK 60
61 DNNNGDDDDE DDDEIVMPVP NVRSSVLQKV IEWAEHHRDS NFPDEDDDDS RKSAPVDSWD 120
121 REFLKVDQEM LYEIILAANY LNIKPLLDAG CKVVAEMIRG RSPEEIRRTF NIVNDFTPEE 180
181 EAAIRRENEW AEDR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
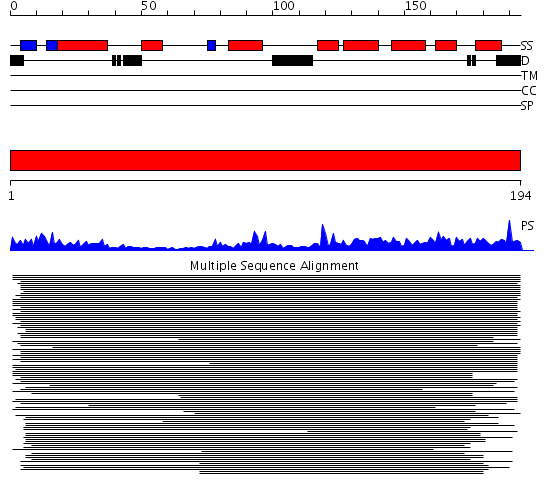
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..194] | 384.68867 | Cyclin A/CDK2-associated p45, Skp1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)