













| General Information: |
|
| Name(s) found: |
CLB1 /
YGR108W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 471 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
G2/M transition of mitotic cell cycle
[IGI mitotic spindle organization in nucleus [IGI meiotic G2/MI transition [IEP regulation of cyclin-dependent protein kinase activity [TAS] |
| Molecular Function: |
cyclin-dependent protein kinase regulator activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRSLLVENS RTINSNEEKG VNESQYILQK RNVPRTILGN VTNNANILQE ISMNRKIGMK 60
61 NFSKLNNFFP LKDDVSRADD FTSSFNDSRQ GVKQEVLNNK ENIPEYGYSE QEKQQCSNDD 120
121 SFHTNSTALS CNRLIYSENK SISTQMEWQK KIMREDSKKK RPISTLVEQD DQKKFKLHEL 180
181 TTEEEVLEEY EWDDLDEEDC DDPLMVSEEV NDIFDYLHHL EIITLPNKAN LYKHKNIKQN 240
241 RDILVNWIIK IHNKFGLLPE TLYLAINIMD RFLCEEVVQL NRLQLVGTSC LFIASKYEEI 300
301 YSPSIKHFAY ETDGACSVED IKEGERFILE KLDFQISFAN PMNFLRRISK ADDYDIQSRT 360
361 LAKFLMEISI VDFKFIGILP SLCASAAMFL SRKMLGKGTW DGNLIHYSGG YTKAKLYPVC 420
421 QLLMDYLVGS TIHDEFLKKY QSRRFLKASI ISIEWALKVR KNGYDIMTLH E |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | McCusker D, et al (2007) | |
| View Run | No Comments | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
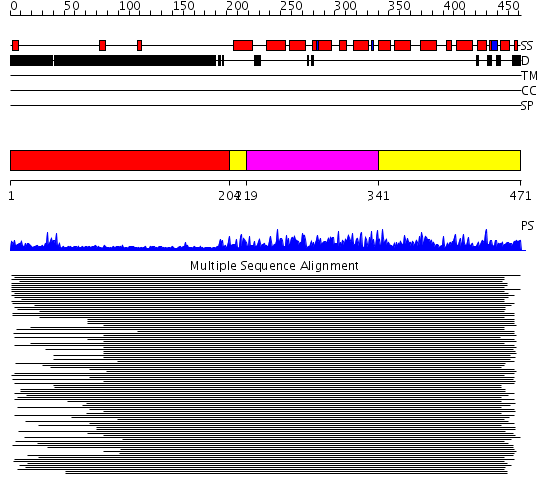
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..203] | 3.29999 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [204..218] [341..471] |
343.9897 | Cyclin A | |
| 3 | View Details | [219..340] | 343.9897 | Cyclin A |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.61 |
Source: Reynolds et al. (2008)