













| General Information: |
|
| Name(s) found: |
RAV1 /
YJR033C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1357 amino acids |
Gene Ontology: |
|
| Cellular Component: |
RAVE complex
[IPI cytoplasm [IDA extrinsic to membrane [IDA |
| Biological Process: |
early endosome to late endosome transport
[IGI regulation of protein complex assembly [IGI vacuolar acidification [IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLNFLPGRP NATPQTACQA TWQNHTIFAY CSGNNLIILT NKFTRLQTIY TQSDCTAVDI 60
61 NSQNGFIALS FHNRVLIYKP IHQIMQNPKW TQCCQLFHDD TPVNCLRWSS DNELAIGSDF 120
121 LSFWKIKDNF GVYQPILQWN QKQPKPVYNV IISQDSQLIV SIGKYDCNAK LWKRVSIVGE 180
181 QAIFNLTMLP HPKPITAMRW KKEPDQVSKN NTASHALYTL CEDKVLRIWS CFEMEKNHTV 240
241 QIWGEVPLSP TQKFCVIIDN WIIRQTLSVK DSEIFDISDS DIVILGSMTG EMEVLALNNL 300
301 SQDPPKPMTK KTISHKKVKK ATMLNDTRYL YLPEIQPYDN VKGKLSFLVH DLQGVIRHLL 360
361 IDILQLINNK TEDLSAALEH KFTGHNKSVQ KLVRSSDGEA LLTTSRFSEN GVWYPQKLNH 420
421 GVSLRLQNTI QTESPIKFAV VHELGKQVIC LLENGALQAW ECPTNRKEDS EQKQSYLRVE 480
481 TRLKEEKKIH PIVMLNTPEP KHSHERHFTA LIFSDGSIKA FEVSLTRGIF EVKSDSLDID 540
541 GDDIYKISII DPVHQTFVSN RPLISLITKK GLTRTYKAIV NYNDRHVQWI KACEINTGIM 600
601 NCTCIRGSST GKLCIVNSTG KVMSLWDLNR GVLEYEETFH NPIEDIDWTS TEYGQSIVSI 660
661 GFTGYALLYT QLRYDYTNNT PSYLPIEKID ITAHTAHNIG DSVWMKNGTF VVASGNQFYI 720
721 KDKSLDLTDP FTYQSIGSRK ILSNDILHLS SVLNGPLPVY HPQFLIQAIY ANKLQLVKEL 780
781 LLRLFLALRK LDFESQDVSN LDSNLGMDPL KYFIAKDRDY PVESFPDPYP CFNKTVSLAL 840
841 TEQLTKTTLP YLTRHQQITL ITVIEAVDEV TKNENIVDYN GVRFLLGVKL FLSHKNIQKS 900
901 ILMRDVSWAL HSDNKEILLS SIDRHITSWN RAREYRIAYW IKEQDLVKKF EDIAKYEFSK 960
961 DDKRDPSRCA IFYLALKKKQ ILLSLWKMAI GHPEQQKMVR FISNDFTVPR WRTAALKNAF1020
1021 VLLSKHRYMD AAVFFLLTDS LKDCVNVLCK QVHDMDLAIG VCRVYEGDNG PVLGELLTAQ1080
1081 MLPETIKEND RWKASFIYWK LRKQEVAIKA LLTAPIDLEN NSSIVDKEVC VNRSFLVEDP1140
1141 ALLYLYNHLR NRNLKYFIGS LNVEAKIECT LILRVTDILC RMGCNYLAVS LVKNWKFIER1200
1201 NSIPVQKLLK SPTKDRAYSA IGAMASEPIS TARMRPSLFD KFGSPSASDI ESPNPKLPNS1260
1261 LLDDFLQPPP NSTSSNSLAQ SSSSAPRSIL DEFVSPSYSQ HKENLTPKAP NDSVGETDNS1320
1321 ENRKDKLSKD ILDDLSSQKP QKPKKSAITK NLLDDFV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
RAV1 VPS72 |
| View Details | Krogan NJ, et al. (2006) |

|
RAV1 RAV2 |
| View Details | Gavin AC, et al. (2002) |
|
RAV1 TFP1 VMA10 VMA13 VMA2 VMA4 VMA5 VMA6 VMA7 VMA8 VPH1 |
| View Details | Qiu et al. (2008) |
|
RAV1 RAV2 SKP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
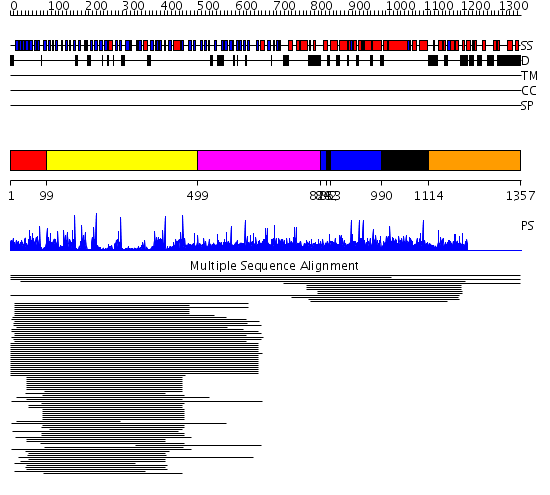
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 38.045757 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 2 | View Details | [99..498] | 42.69897 | Tup1, C-terminal domain | |
| 3 | View Details | [499..827] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [828..841] [853..989] |
9.54 | (+)-bornyl diphosphate synthase | |
| 5 | View Details | [842..852] [990..1113] |
9.54 | (+)-bornyl diphosphate synthase | |
| 6 | View Details | [1114..1357] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1199..1357] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)