













| General Information: |
|
| Name(s) found: |
CMD1 /
YBR109C
[SGD]
|
| Description(s) found:
Found 38 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 147 amino acids |
Gene Ontology: |
|
| Cellular Component: |
incipient cellular bud site
[IDA cellular bud tip [IDA central plaque of spindle pole body [IDA cytoplasm [TAS cellular bud neck [IDA mating projection tip [IDA |
| Biological Process: |
cytoskeleton organization
[IMP cell budding [IMP vacuole fusion, non-autophagic [IMP mitosis [TAS endocytosis [IMP |
| Molecular Function: |
protein binding
[TAS calcium ion binding [IDA calcium-dependent protein binding [TAS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ABP1 BSP1 CMD1 CMP2 CRN1 INP52 LSP1 MLC1 MYO2 MYO3 MYO4 MYO5 NOP53 PIL1 RPL40B, RPL40A RVS161 SAC6 SHE3 YGL242C YNL208W |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
BBP1 CDC31 CMD1 CNM67 KAR1 NUD1 SPC105 SPC110 SPC24 SPC25 SPC29 SPC42 SPC72 |
| View Details | Krogan NJ, et al. (2006) |
|
CMD1 CMK1 CMK2 CMP2 CNA1 CNB1 MYO3 MYO5 YGL242C |
| View Details | Gavin AC, et al. (2002) |

|
CMD1 CSL4 CTR9 DIS3 ECM16 GCN1 MAM33 MLC1 MTR3 MYO2 NMD5 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SAM1 SHE4 SKI6 SKI7 SRP1 UTP10 |
| View Details | Ho Y, et al. (2002) |
|
CMD1 CMK1 CMK2 CMP2 CNA1 COF1 CPR1 EDE1 HCH1 HUL5 HYP2 IDH1 ILS1 IPP1 MLC1 MYO2 MYO3 MYO4 MYO5 PGM2 PST2 RFC3 RPN7 SAY1 SHE3 SHE4 SOD1 SPC110 TEF4 UBA1 VAS1 VPH2 VPS13 YNK1 |
| View Details | Qiu et al. (2008) |
|
CDC31 CMD1 HSK3 KAR1 SPC110 SPC29 SPC42 SPC72 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #33 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #27 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps3-5 | Keck JM, et al. (2011) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #35 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #01 Alpha factor Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #28 Asynchronous Prep5-TiO2 Phosphopeptide Enrichment | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #15 Mitotic 2nd-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
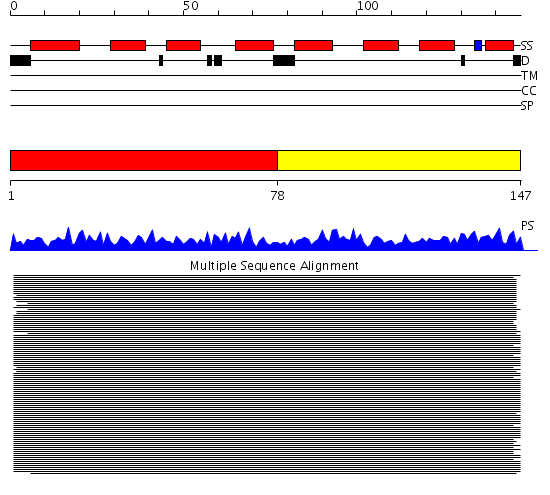
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | 481.67837 | Calmodulin | |
| 2 | View Details | [78..147] | 481.67837 | Calmodulin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)