













| General Information: |
|
| Name(s) found: |
TRF5 /
YNL299W
[SGD]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 625 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS nucleolus [IDA |
| Biological Process: |
ncRNA polyadenylation
[IGI mitotic sister chromatid cohesion [IGI rRNA catabolic process [IGI snRNA catabolic process [IGI snoRNA catabolic process [IGI ncRNA polyadenylation involved in polyadenylation-dependent ncRNA catabolic process [IGI |
| Molecular Function: |
DNA-directed DNA polymerase activity
[IDA polynucleotide adenylyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTRLKAKYSP TKGKRKEDKH TKRMRKSSFT RTQKMLEVFN DNRSHFNKYE SLAIDVEDDD 60
61 TFGNLVLMEN DKSDVDIPVI EEVTSSEDEQ RAESSKRNNS LEDNQDFIAF SDSSEDETEQ 120
121 IKEDDDERSS FLLTDEHEVS KLTSQQSLNT ESACNVEYPW IRNHCHSKQR RIADWLTSEI 180
181 KDFVHYISPS KNEIKCRNRT IDKLRRAVKE LWSDADLHVF GSFATDLYLP GSDIDCVVNS 240
241 RNRDKEDRNY IYELARHLKN KGLAIRMEVI VKTRVPIIKF IEPQSQLHID VSFERTNGLE 300
301 AAKLIREWLR DSPGLRELVL IIKQFLHSRR LNNVHTGGLG GFTVICLVYS FLNMHPRIKS 360
361 NDIDVLDNLG VLLIDFFELY GKNFGYDDVA ISISDGYASY IPKSCWRTLE PSRSKFSLAI 420
421 QDPGDPNNNI SRGSFNMKDI KKAFAGAFEL LVNKCWELNS ATFKDRVGKS ILGNVIKYRG 480
481 QKRDFNDERD LVQNKAIIEN ERYHKRRTRI VQEDLFINDT EDLPVEEIYK LDEPAKKKQK 540
541 AKKDKREGEI KKSAIPSPPP DFGVSRSKLK RKVKKTDQGS LLHQNNLSID DLMGLSENDQ 600
601 ESDQDQKGRD TLRDKMRNHH WKLRQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
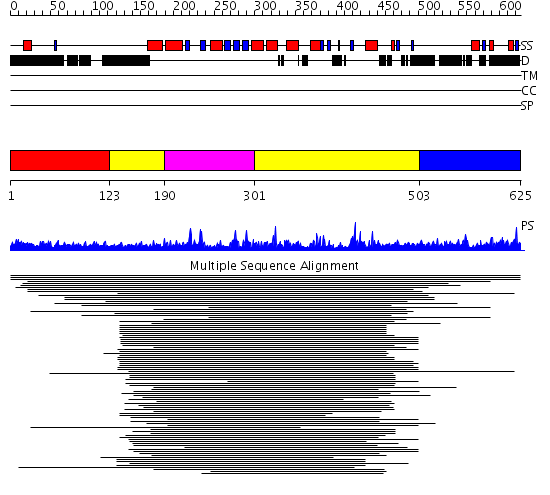
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [123..189] [301..502] |
65.09691 | Poly(A) polymerase, middle domain; Poly(A) polymerase N-terminal, catalytic domain; Poly(A) polymerase, C-terminal domain | |
| 3 | View Details | [190..300] | 65.09691 | Poly(A) polymerase, middle domain; Poly(A) polymerase N-terminal, catalytic domain; Poly(A) polymerase, C-terminal domain | |
| 4 | View Details | [503..625] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.49 |
Source: Reynolds et al. (2008)