













| General Information: |
|
| Name(s) found: |
URB1 /
YKL014C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1764 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
rRNA processing
[IMP processing of 27S pre-rRNA [IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNHSEAYGS RDQRREKYTQ GKEFEDGTLE TLESIISAVE DETLSKDYQP LIVFFQRGFG 60
61 AQLVQTWSYY AQVNNHGKFS KTTSLLTKTL RVLSSDTSTV TIGSGLIRLI LTDYTKVLYR 120
121 GLNNMRAQLT NPILRLLKQI VNFNNGQHIE ELVSYFDFSL PILPRLLVPS KSELANGNSS 180
181 ADSSKHDSLR FTFIKFWLTL ISNASPFVRK ELLTENFKIM SNLFKFMNKA DSDKLSEHIL 240
241 SVFINDILKE KSFKRTTKTK ILNELAASKI HHFYYSSNKN LVKKANEFFL TFGASRDFSV 300
301 AFPDNCVWFK NSVADGASHG APITVNQVEF QIHNKLLFNT LRLFKPWEDT LQLGTLIKIL 360
361 ENVPELVAPY SIFLTTNGNY DPKMTSYWFG ITLLINKIIN LKIPQFMEKV DSNIPPATSL 420
421 VIENILPSLL TKSSLVKSLQ FETPIIRQLA CQSIVLALKK LEKVSTFYDQ KGWRNEKTIL 480
481 LNEFHTRIPD LPIFVSTLSN SLASNKDNRI LPLSISIIFN YYSKMFPNLF SINLPSSNIY 540
541 TDIMQKSKIS GIEFAILDNY LQFQEFNSTQ TRWWNPSSGG NSLFTLLLKL ASSKNASNVI 600
601 TTRISNLLDE LTRTNVIFNI SLISPVMALV NSLQGLSLQV SEIDNMEQVW KWFDETISRV 660
661 VKTPYKYVDM AKEYNYISPF IMCLSEQWKY VDKSGNPEFL IKWLILFLRN MIFIGEDHIG 720
721 IDKLVKNVFP EVSDHDVNIY LKLDSFEENI KKTNSSNSLI SSMKSSSFFQ YISALPSKNL 780
781 MNISRLPVNK LDAAGILFRV QLLVEDDSVV YDNWFEATAC ELTGKIASYM VTDTEFPIIK 840
841 VLERYINFAL PKLAIEKRNA LLMKKSRFMC NLIGAVCFET GHQLVEFREI IQKVVFSGEN 900
901 VEEYANYNEL YQKEDVNAFL TSVSEYLSTS ALTSLLMCST KLESTRNILQ KLFNEGKTIK 960
961 ISLVKNILNK AANEDPASIK EVNISLAKFF EENKVCVDAS SDPMGKLSLS ETTSLINSFV1020
1021 SSDLNYLVLK AFYRWEHFSF PSFIPSIWRI KDSPLLSIVT TAALFKHMQD KDFSAFAHET1080
1081 ISKYGNEIAK STYTTSKSEI FDEILNMITT YIDFYDETKR NEILKCVLSQ SDHKYHAATV1140
1141 RYIAAHNNFT YPGVETWLHK TLLYLTKYLS ERKVISNSFF ELLRAMAELL KLEEVPNKLN1200
1201 VKIINSQLEA ILGSEWIKQI KVLEYVIVLI FCVSKKSIQS QRMVQLLLSN DSYSSIMIKD1260
1261 NDEDSSYRKF LSTMILFSLF SIDPVVNSTP IVQEKLLTFY SGTISSNDKL ILKILETIES1320
1321 HTATSWTNMI FSWEFIKDEE EEILEAIGDT RLITKEREGL ILTLQKNMIK KSIDRYVLER1380
1381 PQVPELYTDS NTNNYDATTR CDLVKKYYDD TERSGVDMYD PLFLLLLIIH NKELVKMVKD1440
1441 DEGNVTYRYE FENFLDCKIF QFIICSLSDC HTVANISYEH LSNLASSLEK KTAQMNLEKQ1500
1501 ITSKDNERKE SDSDLIKYNS IYQVLIKRIL YQRQQNQDPI NPLIWFSISR IVDLLGSPTA1560
1561 PLHEKAYRWV LSNSTIRSWD IPMVSDVMMS YNKRQQDDNK KEIDMEIYYG ELSWVLTTIC1620
1621 KGIKTDEDYK MLEKKGVFEW LLNLINMPYL KERLRELIYF IFYKVQRVAD DGGLNLISRN1680
1681 GIVSFFEVLN NNIKSRLPQD DILNNIGTLR NENRGTLNTT LRLAQEQNGI EKLLLGYNEL1740
1741 VKSQKRLILW TEGDSDNVVK RLRK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
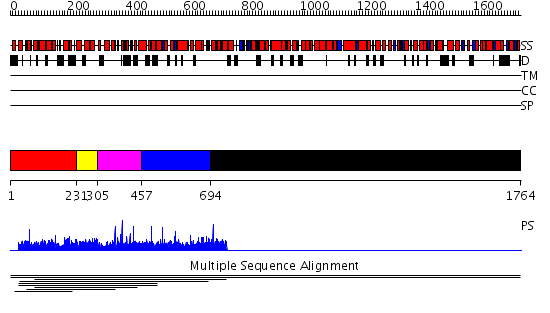
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..230] | 2.59 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 2 | View Details | [231..304] | 2.59 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 3 | View Details | [305..456] | 2.59 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [457..693] | 2.59 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 5 | View Details | [694..1764] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1288..1511] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1512..1764] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)