













| General Information: |
|
| Name(s) found: |
SRP1 /
YNL189W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 542 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
nucleocytoplasmic transport
[TAS |
| Molecular Function: |
protein transmembrane transporter activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNGTDSSTS KFVPEYRRTN FKNKGRFSAD ELRRRRDTQQ VELRKAKRDE ALAKRRNFIP 60
61 PTDGADSDEE DESSVSADQQ FYSQLQQELP QMTQQLNSDD MQEQLSATVK FRQILSREHR 120
121 PPIDVVIQAG VVPRLVEFMR ENQPEMLQLE AAWALTNIAS GTSAQTKVVV DADAVPLFIQ 180
181 LLYTGSVEVK EQAIWALGNV AGDSTDYRDY VLQCNAMEPI LGLFNSNKPS LIRTATWTLS 240
241 NLCRGKKPQP DWSVVSQALP TLAKLIYSMD TETLVDACWA ISYLSDGPQE AIQAVIDVRI 300
301 PKRLVELLSH ESTLVQTPAL RAVGNIVTGN DLQTQVVINA GVLPALRLLL SSPKENIKKE 360
361 ACWTISNITA GNTEQIQAVI DANLIPPLVK LLEVAEYKTK KEACWAISNA SSGGLQRPDI 420
421 IRYLVSQGCI KPLCDLLEIA DNRIIEVTLD ALENILKMGE ADKEARGLNI NENADFIEKA 480
481 GGMEKIFNCQ QNENDKIYEK AYKIIETYFG EEEDAVDETM APQNAGNTFG FGSNVNQQFN 540
541 FN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
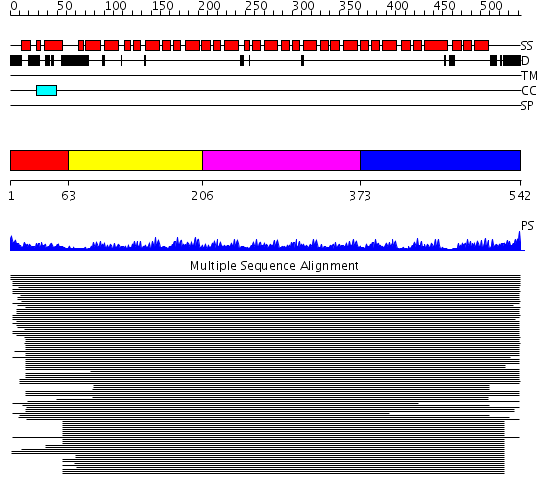
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..62] | 43.69897 | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA | |
| 2 | View Details | [63..205] | 10107.0 | Karyopherin alpha | |
| 3 | View Details | [206..372] | 10107.0 | Karyopherin alpha | |
| 4 | View Details | [373..542] | 10107.0 | Karyopherin alpha |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)