













| General Information: |
|
| Name(s) found: |
TFP1 /
YDL185W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1071 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar proton-transporting V-type ATPase, V1 domain
[TAS fungal-type vacuole membrane [IDA |
| Biological Process: |
protein metabolic process
[IDA vacuolar acidification [TAS intron homing [TAS |
| Molecular Function: |
proton-transporting ATPase activity, rotational mechanism
[TAS endodeoxyribonuclease activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGAIENARK EIKRISLEDH AESEYGAIYS VSGPVVIAEN MIGCAMYELV KVGHDNLVGE 60
61 VIRIDGDKAT IQVYEETAGL TVGDPVLRTG KPLSVELGPG LMETIYDGIQ RPLKAIKEES 120
121 QSIYIPRGID TPALDRTIKW QFTPGKFQVG DHISGGDIYG SVFENSLISS HKILLPPRSR 180
181 GTITWIAPAG EYTLDEKILE VEFDGKKSDF TLYHTWPVRV PRPVTEKLSA DYPLLTGQRV 240
241 LDALFPCVQG GTTCIPGAFG CGKTVISQSL SKYSNSDAII YVGCFAKGTN VLMADGSIEC 300
301 IENIEVGNKV MGKDGRPREV IKLPRGRETM YSVVQKSQHR AHKSDSSREV PELLKFTCNA 360
361 THELVVRTPR SVRRLSRTIK GVEYFEVITF EMGQKKAPDG RIVELVKEVS KSYPISEGPE 420
421 RANELVESYR KASNKAYFEW TIEARDLSLL GSHVRKATYQ TYAPILYEND HFFDYMQKSK 480
481 FHLTIEGPKV LAYLLGLWIG DGLSDRATFS VDSRDTSLME RVTEYAEKLN LCAEYKDRKE 540
541 PQVAKTVNLY SKVVRGNGIR NNLNTENPLW DAIVGLGFLK DGVKNIPSFL STDNIGTRET 600
601 FLAGLIDSDG YVTDEHGIKA TIKTIHTSVR DGLVSLARSL GLVVSVNAEP AKVDMNGTKH 660
661 KISYAIYMSG GDVLLNVLSK CAGSKKFRPA PAAAFARECR GFYFELQELK EDDYYGITLS 720
721 DDSDHQFLLA NQVVVHNCGE RGNEMAEVLM EFPELYTEMS GTKEPIMKRT TLVANTSNMP 780
781 VAAREASIYT GITLAEYFRD QGKNVSMIAD SSSRWAEALR EISGRLGEMP ADQGFPAYLG 840
841 AKLASFYERA GKAVALGSPD RTGSVSIVAA VSPAGGDFSD PVTTATLGIT QVFWGLDKKL 900
901 AQRKHFPSIN TSVSYSKYTN VLNKFYDSNY PEFPVLRDRM KEILSNAEEL EQVVQLVGKS 960
961 ALSDSDKITL DVATLIKEDF LQQNGYSTYD AFCPIWKTFD MMRAFISYHD EAQKAVANGA1020
1021 NWSKLADSTG DVKHAVSSSK FFEPSRGEKE VHGEFEKLLS TMQERFAEST D |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
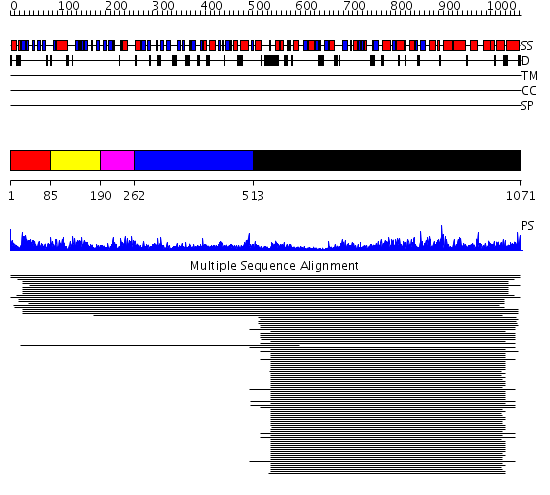
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 21.886057 | No description for PF02874.7 was found. No confident structure predictions are available. | |
| 2 | View Details | [85..189] | 2.05899 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [190..261] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [262..512] | 80.0 | VMA1-derived endonuclease (VDE) PI-Scei intein; VMA1-derived endonuclease (VDE) PI-SceI | |
| 5 | View Details | [513..1071] | 113.0 | F1 ATP synthase alpha subunit, domain 3; F1 ATP synthase alpha subunit, domain 1; Central domain of alpha subunit of F1 ATP synthase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)