













| General Information: |
|
| Name(s) found: |
HEF3 /
YNL014W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1044 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosolic ribosome
[IPI]
|
| Biological Process: |
translational elongation
[IGI |
| Molecular Function: |
ATPase activity
[IDA translation elongation factor activity [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDSQQSITV LEELFRKLET ATSETREGIS SELSSFLNGN IIEHDVPEVF FDEFQKAIQS 60
61 KQKALNTLGA VAYIANETNL SPSVEPYIVA TVPSVCSKAG SKDNDVQLAA TKALKAIASA 120
121 VNPVAVKALL PHLIHSLETS NKWKEKVAVL EVISVLVDAA KEQIALRMPE LIPVLSESMW 180
181 DTKKGVKEAA TTTITKATET VDNKDIERFI PKLIECIANP NEVPETVHLL GATTFVAEVT 240
241 PATLSIMVPL LSRGLAERET SIKRKAAVII DNMCKLVEDP QVVAPFLGKL LPGLKNNFAT 300
301 IADPEAREVT LKALKTLRRV GNVGEDDVLP EISHAGDVST TLGVIKELLE PEKVAPRFTI 360
361 VVEYIAAIAA NLIDERIIDQ QTWFTHVTPY MTIFLHEKTA KEILDDFRKR AVDNIPVGPN 420
421 FQDEEDEGED LCNCEFSLAY GAKILLNKTQ LRLKRGRRYG LCGPNGAGKS TLMRSIANGQ 480
481 VDGFPTQDEC RTVYVEHDID NTHSDMSVLD FVYSGNVGTK DVITSKLKEF GFSDEMIEMP 540
541 IASLSGGWKM KLALARAVLK DADILLLDEP TNHLDTVNVE WLVNYLNTCG ITSVIVSHDS 600
601 GFLDKVCQYI IHYEGLKLRK YKGNLSEFVQ KCPTAQSYYE LGASDLEFQF PTPGYLEGVK 660
661 TKQKAIVKVS NMTFQYPGTT KPQVSDVTFQ CSLSSRIAVI GPNGAGKSTL INVLTGELLP 720
721 TSGEVYTHEN CRIAYIKQHA FAHIESHLDK TPSEYIQWRF QTGEDRETMD RANRQINEND 780
781 AEAMNKIFKI EGTPRRVAGI HSRRKFKNTY EYECSFLLGE NIGMKSERWV PMMSVDNAWL 840
841 PRGELIESHS KMVAEIDMKE ALASGQFRAL TRKEIELHCA MLGLDSELVS HSRIRGLSGG 900
901 QKVKLVLAAC TWQRPHLIVL DEPTNYLDRD SLGALSKALK AFEGGVIIIT HSAEFTKNLT 960
961 DEVWAVKDGK MTPSGHNWVA GQGAGPRIEK KEEEGDKFDA MGNKINSGKK KSKLSSAELR1020
1021 KKKKERMKKK KEMGDEYVSS DEDF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |

|
AAC3 AAT2 ACH1 ACO1 ADE5,7 ADK1 AFR1 AHA1 AHP1 ARC1 ARF1 ARO1 ATP3 ATP5 ATP7 BCY1 BUD32 CAR2 CDC14 CDC15 CDC33 CDC60 CLU1 COF1 COR1 CPA2 CPR1 CPR6 CRM1 CYR1 CYS4 DBP8 DEF1 DIA4 DPM1 EGD1 ERG13 ERG6 ESS1 FAA1 FPR1 FRS2 FUR1 GDE1 GLC7 GND1 GND2 GPG1 GRX1 GRX3 GRX4 GUS1 HEF3 HHF1, HHF2 HMS1 HOR2 HSP104 HTB1 HXT6 HXT7 IDP2 IMD2 IMD4 KAE1 KGD1 KIP3 LHS1 LRO1 LSP1 MCR1 MDH1 MDN1 MET6 MGE1 MGT1 MTC5 NPA3 NTF2 OLA1 OYE2 PFK1 PGM2 PHO81 PIL1 PRM2 PRO2 PTC6 PUP2 PYC1 RAD1 RFA3 RFC2 RHR2 RIP1 RNH202 RNR1 RNR2 RPB3 RPN1 RPN5 RPN6 RPO21 RPT1 RSN1 SAH1 SCP160 SEC18 SEC23 SEC26 SEC27 SEC53 SEN15 SES1 SGO1 SNF4 SNU13 SOD1 SOF1 SPE3 SPT5 SRP1 SSZ1 TEF4 TFG1 THI22 THS1 TIF1, TIF2 TMA19 TMA29 TOM1 TPS1 TSA2 UBA1 UBR1 URA7 UTP22 VAS1 VMA4 VMA5 WTM1 YDL086W YHR033W YJR141W YNK1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #28 Asynchronous Prep5-TiO2 Phosphopeptide Enrichment | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
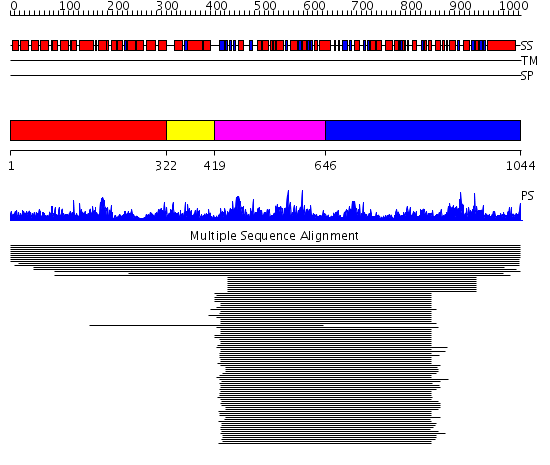
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..321] | 6.11895 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [322..418] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [419..645] | 93.383689 | ATP-binding subunit of the histidine permease | |
| 4 | View Details | [646..1044] | 7.425969 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)