













| General Information: |
|
| Name(s) found: |
FKS1 /
YLR342W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1876 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA 1,3-beta-glucan synthase complex [TAS actin cortical patch [IDA actin cap [IDA |
| Biological Process: |
1,3-beta-glucan biosynthetic process
[TAS regulation of cell size [IMP cellular cell wall organization [TAS endocytosis [IMP |
| Molecular Function: |
1,3-beta-glucan synthase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNTDQQPYQG QTDYTQGPGN GQSQEQDYDQ YGQPLYPSQA DGYYDPNVAA GTEADMYGQQ 60
61 PPNESYDQDY TNGEYYGQPP NMAAQDGENF SDFSSYGPPG TPGYDSYGGQ YTASQMSYGE 120
121 PNSSGTSTPI YGNYDPNAIA MALPNEPYPA WTADSQSPVS IEQIEDIFID LTNRLGFQRD 180
181 SMRNMFDHFM VLLDSRSSRM SPDQALLSLH ADYIGGDTAN YKKWYFAAQL DMDDEIGFRN 240
241 MSLGKLSRKA RKAKKKNKKA MEEANPEDTE ETLNKIEGDN SLEAADFRWK AKMNQLSPLE 300
301 RVRHIALYLL CWGEANQVRF TAECLCFIYK CALDYLDSPL CQQRQEPMPE GDFLNRVITP 360
361 IYHFIRNQVY EIVDGRFVKR ERDHNKIVGY DDLNQLFWYP EGIAKIVLED GTKLIELPLE 420
421 ERYLRLGDVV WDDVFFKTYK ETRTWLHLVT NFNRIWVMHI SIFWMYFAYN SPTFYTHNYQ 480
481 QLVDNQPLAA YKWASCALGG TVASLIQIVA TLCEWSFVPR KWAGAQHLSR RFWFLCIIFG 540
541 INLGPIIFVF AYDKDTVYST AAHVVAAVMF FVAVATIIFF SIMPLGGLFT SYMKKSTRRY 600
601 VASQTFTAAF APLHGLDRWM SYLVWVTVFA AKYSESYYFL VLSLRDPIRI LSTTAMRCTG 660
661 EYWWGAVLCK VQPKIVLGLV IATDFILFFL DTYLWYIIVN TIFSVGKSFY LGISILTPWR 720
721 NIFTRLPKRI YSKILATTDM EIKYKPKVLI SQVWNAIIIS MYREHLLAID HVQKLLYHQV 780
781 PSEIEGKRTL RAPTFFVSQD DNNFETEFFP RDSEAERRIS FFAQSLSTPI PEPLPVDNMP 840
841 TFTVLTPHYA ERILLSLREI IREDDQFSRV TLLEYLKQLH PVEWECFVKD TKILAEETAA 900
901 YEGNENEAEK EDALKSQIDD LPFYCIGFKS AAPEYTLRTR IWASLRSQTL YRTISGFMNY 960
961 SRAIKLLYRV ENPEIVQMFG GNAEGLEREL EKMARRKFKF LVSMQRLAKF KPHELENAEF1020
1021 LLRAYPDLQI AYLDEEPPLT EGEEPRIYSA LIDGHCEILD NGRRRPKFRV QLSGNPILGD1080
1081 GKSDNQNHAL IFYRGEYIQL IDANQDNYLE ECLKIRSVLA EFEELNVEQV NPYAPGLRYE1140
1141 EQTTNHPVAI VGAREYIFSE NSGVLGDVAA GKEQTFGTLF ARTLSQIGGK LHYGHPDFIN1200
1201 ATFMTTRGGV SKAQKGLHLN EDIYAGMNAM LRGGRIKHCE YYQCGKGRDL GFGTILNFTT1260
1261 KIGAGMGEQM LSREYYYLGT QLPVDRFLTF YYAHPGFHLN NLFIQLSLQM FMLTLVNLSS1320
1321 LAHESIMCIY DRNKPKTDVL VPIGCYNFQP AVDWVRRYTL SIFIVFWIAF VPIVVQELIE1380
1381 RGLWKATQRF FCHLLSLSPM FEVFAGQIYS SALLSDLAIG GARYISTGRG FATSRIPFSI1440
1441 LYSRFAGSAI YMGARSMLML LFGTVAHWQA PLLWFWASLS SLIFAPFVFN PHQFAWEDFF1500
1501 LDYRDYIRWL SRGNNQYHRN SWIGYVRMSR ARITGFKRKL VGDESEKAAG DASRAHRTNL1560
1561 IMAEIIPCAI YAAGCFIAFT FINAQTGVKT TDDDRVNSVL RIIICTLAPI AVNLGVLFFC1620
1621 MGMSCCSGPL FGMCCKKTGS VMAGIAHGVA VIVHIAFFIV MWVLESFNFV RMLIGVVTCI1680
1681 QCQRLIFHCM TALMLTREFK NDHANTAFWT GKWYGKGMGY MAWTQPSREL TAKVIELSEF1740
1741 AADFVLGHVI LICQLPLIII PKIDKFHSIM LFWLKPSRQI RPPIYSLKQT RLRKRMVKKY1800
1801 CSLYFLVLAI FAGCIIGPAV ASAKIHKHIG DSLDGVVHNL FQPINTTNND TGSQMSTYQS1860
1861 HYYTHTPSLK TWSTIK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ANP1 BET5 FKS1 GEA2 MIR1 MNN10 PHB2 RPL17A RPL24A RPL28 RPS17B SEC7 TCB3 TRS23 YHR020W |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
FEN1 FKS1 GSC2 |
| View Details | Krogan NJ, et al. (2006) |

|
CTP1 ERV25 FKS1 FTR1 MRH1 OLE1 PEX27 ROG3 RPL10 RPL36B RPL37A RPP1A SEC11 SPC1 SPC2 SPC3 TCM62 TSC13 YNR021W |
| View Details | Gavin AC, et al. (2002) |

|
ADH1 ANP1 ARO1 ARX1 BET3 BET5 BMH1 BMH2 CCT4 CLU1 CRM1 CSE1 DHH1 DRS2 DUG1 ECM29 EMP24 ERO1 ERP1 ERP2 ERV25 ERV41 ERV46 FET5 FKS1 FLC2 FTH1 GCD6 GCN1 GCN20 GEA2 GFA1 GSG1 HAS1 HHF1, HHF2 HRP1 IDH2 KAP104 KAP123 KRE11 KTR3 LCB1 LCB2 LYS12 MIR1 MNN10 NAB2 NAT1 NEM1 NIP7 NUM1 OPI1 OSH2 PDR5 PHB1 PIL1 RGR1 RPN10 RPN3 RRP1 RVB2 SAM1 SAM2 SCS2 SEC13 SEC62 SEC63 SEC66 SEC7 SEC72 SKN1 SLC1 SRV2 STT4 SWH1 SWI3 TCB3 TRS120 TRS130 TRS20 TRS23 TRS31 TRS33 YAR044W YHR020W |
| View Details | Qiu et al. (2008) |
|
FKS1 GSC2 RHO1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #01 Alpha factor Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #28 Asynchronous Prep5-TiO2 Phosphopeptide Enrichment | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #09 Alpha Factor Prep4-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Lockshon D, et al. (2006) | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
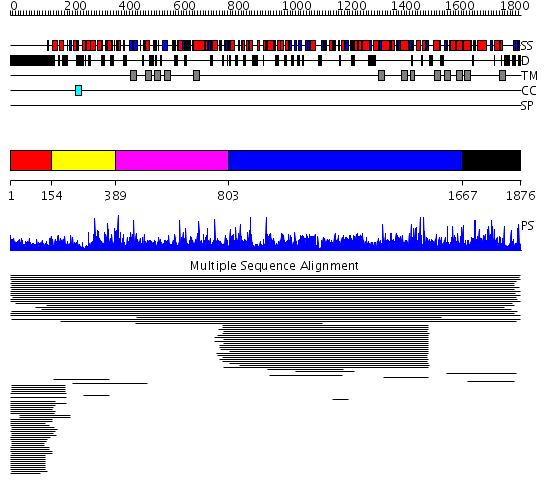
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..153] | 108.145997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [154..388] | 1.05194 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [389..802] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [803..1666] | 1000.0 | 1,3-beta-glucan synthase component No confident structure predictions are available. | |
| 5 | View Details | [1667..1876] | 1.019943 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [620..784] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [785..850] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [851..941] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [942..1106] | 2.048945 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1107..1237] | 3.048945 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1238..1440] | 2.050926 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1441..1542] | 1.046984 | View MSA. No confident structure predictions are available. | |
| 13 | View Details | [1543..1666] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [1667..1876] | 2.023941 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|